Next-Generation Sequencing for Binary Protein-Protein Interactions
- PMID: 26734059
- PMCID: PMC4681833
- DOI: 10.3389/fgene.2015.00346
Next-Generation Sequencing for Binary Protein-Protein Interactions
Abstract
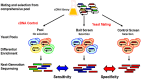
The yeast two-hybrid (Y2H) system exploits host cell genetics in order to display binary protein-protein interactions (PPIs) via defined and selectable phenotypes. Numerous improvements have been made to this method, adapting the screening principle for diverse applications, including drug discovery and the scale-up for proteome wide interaction screens in human and other organisms. Here we discuss a systematic workflow and analysis scheme for screening data generated by Y2H and related assays that includes high-throughput selection procedures, readout of comprehensive results via next-generation sequencing (NGS), and the interpretation of interaction data via quantitative statistics. The novel assays and tools will serve the broader scientific community to harness the power of NGS technology to address PPI networks in health and disease. We discuss examples of how this next-generation platform can be applied to address specific questions in diverse fields of biology and medicine.
Keywords: interactome mapping; next-generation sequencing; protein–protein interactions; quantitative interaction profiles; yeast two-hybrid.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

