Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer
- PMID: 26735014
- PMCID: PMC4854653
- DOI: 10.1038/nature16508
Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer
Abstract
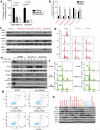
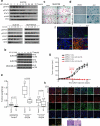
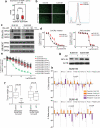
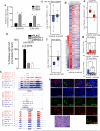
Triple-negative breast cancer (TNBC) is a heterogeneous and clinically aggressive disease for which there is no targeted therapy. BET bromodomain inhibitors, which have shown efficacy in several models of cancer, have not been evaluated in TNBC. These inhibitors displace BET bromodomain proteins such as BRD4 from chromatin by competing with their acetyl-lysine recognition modules, leading to inhibition of oncogenic transcriptional programs. Here we report the preferential sensitivity of TNBCs to BET bromodomain inhibition in vitro and in vivo, establishing a rationale for clinical investigation and further motivation to understand mechanisms of resistance. In paired cell lines selected for acquired resistance to BET inhibition from previously sensitive TNBCs, we failed to identify gatekeeper mutations, new driver events or drug pump activation. BET-resistant TNBC cells remain dependent on wild-type BRD4, which supports transcription and cell proliferation in a bromodomain-independent manner. Proteomic studies of resistant TNBC identify strong association with MED1 and hyper-phosphorylation of BRD4 attributable to decreased activity of PP2A, identified here as a principal BRD4 serine phosphatase. Together, these studies provide a rationale for BET inhibition in TNBC and present mechanism-based combination strategies to anticipate clinical drug resistance.
Figures





Comment in
-
Cancer: Bet on drug resistance.Nature. 2016 Jan 21;529(7586):289-90. doi: 10.1038/nature16863. Epub 2016 Jan 6. Nature. 2016. PMID: 26735017 No abstract available.
References
-
- Metzger-Filho O, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30:1879–1887. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

