FOXO1 couples metabolic activity and growth state in the vascular endothelium
- PMID: 26735015
- PMCID: PMC5380221
- DOI: 10.1038/nature16498
FOXO1 couples metabolic activity and growth state in the vascular endothelium
Abstract
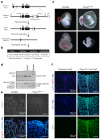
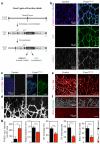
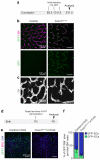
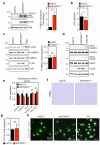
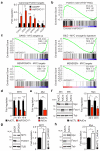
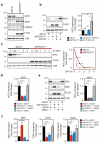
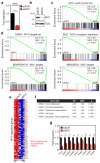
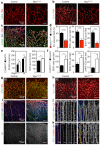
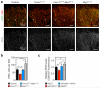
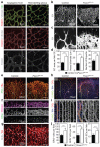
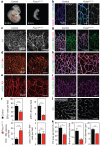
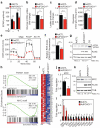
Endothelial cells (ECs) are plastic cells that can switch between growth states with different bioenergetic and biosynthetic requirements. Although quiescent in most healthy tissues, ECs divide and migrate rapidly upon proangiogenic stimulation. Adjusting endothelial metabolism to the growth state is central to normal vessel growth and function, yet it is poorly understood at the molecular level. Here we report that the forkhead box O (FOXO) transcription factor FOXO1 is an essential regulator of vascular growth that couples metabolic and proliferative activities in ECs. Endothelial-restricted deletion of FOXO1 in mice induces a profound increase in EC proliferation that interferes with coordinated sprouting, thereby causing hyperplasia and vessel enlargement. Conversely, forced expression of FOXO1 restricts vascular expansion and leads to vessel thinning and hypobranching. We find that FOXO1 acts as a gatekeeper of endothelial quiescence, which decelerates metabolic activity by reducing glycolysis and mitochondrial respiration. Mechanistically, FOXO1 suppresses signalling by MYC (also known as c-MYC), a powerful driver of anabolic metabolism and growth. MYC ablation impairs glycolysis, mitochondrial function and proliferation of ECs while its EC-specific overexpression fuels these processes. Moreover, restoration of MYC signalling in FOXO1-overexpressing endothelium normalizes metabolic activity and branching behaviour. Our findings identify FOXO1 as a critical rheostat of vascular expansion and define the FOXO1-MYC transcriptional network as a novel metabolic checkpoint during endothelial growth and proliferation.
Figures

Comment in
-
Vascular biology: Transcriptional control of endothelial energy.Nature. 2016 Jan 14;529(7585):160-1. doi: 10.1038/nature16866. Epub 2016 Jan 6. Nature. 2016. PMID: 26735011 No abstract available.
References
-
- Ghesquiere B, Wong BW, Kuchnio A, Carmeliet P. Metabolism of stromal and immune cells in health and disease. Nature. 2014;511:167–176. - PubMed
-
- Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–478. - PubMed
-
- Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–887. - PubMed
-
- De Bock K, Georgiadou M, Carmeliet P. Role of endothelial cell metabolism in vessel sprouting. Cell Metabolism. 2013;18:634–647. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

