Molecular Evolution of the Nuclear Factor (Erythroid-Derived 2)-Like 2 Gene Nrf2 in Old World Fruit Bats (Chiroptera: Pteropodidae)
- PMID: 26735303
- PMCID: PMC4703304
- DOI: 10.1371/journal.pone.0146274
Molecular Evolution of the Nuclear Factor (Erythroid-Derived 2)-Like 2 Gene Nrf2 in Old World Fruit Bats (Chiroptera: Pteropodidae)
Abstract
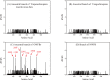
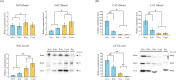
Mammals developed antioxidant systems to defend against oxidative damage in their daily life. Enzymatic antioxidants and low molecular weight antioxidants (LMWAs) constitute major parts of the antioxidant systems. Nuclear factor (erythroid-derived 2)-like 2 (Nrf2, encoded by the Nrf2 gene) is a central transcriptional regulator, regulating transcription, of many antioxidant enzymes. Frugivorous bats eat large amounts of fruits that contain high levels of LMWAs such as vitamin C, thus, a reliance on LMWAs might greatly reduce the need for antioxidant enzymes in comparison to insectivorous bats. Therefore, it is possible that frugivorous bats have a reduced need for Nrf2 function due to their substantial intake of diet-antioxidants. To test whether the Nrf2 gene has undergone relaxed evolution in fruit-eating bats, we obtained Nrf2 sequences from 16 species of bats, including four Old World fruit bats (Pteropodidae) and one New World fruit bat (Phyllostomidae). Our molecular evolutionary analyses revealed changes in the selection pressure acting on Nrf2 gene and identified seven specific amino acid substitutions that occurred on the ancestral lineage leading to Old World fruit bats. Biochemical experiments were conducted to examine Nrf2 in Old World fruit bats and showed that the amount of catalase, which is regulated by Nrf2, was significantly lower in the brain, heart and liver of Old World fruit bats despite higher levels of Nrf2 protein in Old World fruit bats. Computational predictions suggest that three of these seven amino acid replacements might be deleterious to Nrf2 function. Therefore, the results suggest that Nrf2 gene might have experienced relaxed constraint in Old World fruit bats, however, we cannot rule out the possibility of positive selection. Our study provides the first data on the molecular adaptation of Nrf2 gene in frugivorous bats in compensation to the increased levels of LWMAs from their fruit-diet.
Conflict of interest statement
Figures


Similar articles
-
Relaxed evolution in the tyrosine aminotransferase gene tat in old world fruit bats (Chiroptera: Pteropodidae).PLoS One. 2014 May 13;9(5):e97483. doi: 10.1371/journal.pone.0097483. eCollection 2014. PLoS One. 2014. PMID: 24824435 Free PMC article.
-
Adaptive evolution in the glucose transporter 4 gene Slc2a4 in Old World fruit bats (family: Pteropodidae).PLoS One. 2012;7(4):e33197. doi: 10.1371/journal.pone.0033197. Epub 2012 Apr 6. PLoS One. 2012. PMID: 22493665 Free PMC article.
-
Phosphoenolpyruvate carboxykinase 1 gene (Pck1) displays parallel evolution between Old World and New World fruit bats.PLoS One. 2015 Mar 25;10(3):e0118666. doi: 10.1371/journal.pone.0118666. eCollection 2015. PLoS One. 2015. PMID: 25807515 Free PMC article.
-
Simultaneous Activation of Nrf2 and Elevation of Dietary and Endogenous Antioxidant Chemicals for Cancer Prevention in Humans.J Am Coll Nutr. 2016;35(2):175-84. doi: 10.1080/07315724.2014.1003419. Epub 2015 Jul 7. J Am Coll Nutr. 2016. PMID: 26151600 Review.
-
Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation.Mol Aspects Med. 2011 Aug;32(4-6):234-46. doi: 10.1016/j.mam.2011.10.006. Epub 2011 Oct 15. Mol Aspects Med. 2011. PMID: 22020111 Review.
Cited by
-
High basal heat-shock protein expression in bats confers resistance to cellular heat/oxidative stress.Cell Stress Chaperones. 2019 Jul;24(4):835-849. doi: 10.1007/s12192-019-01013-y. Epub 2019 Jun 22. Cell Stress Chaperones. 2019. PMID: 31230214 Free PMC article.
-
Biomimetics provides lessons from nature for contemporary ways to improve human health.J Clin Transl Sci. 2021 May 17;5(1):e128. doi: 10.1017/cts.2021.790. eCollection 2021. J Clin Transl Sci. 2021. PMID: 34367673 Free PMC article. Review.
-
Manipulating the exposome to enable better ageing.Biochem J. 2021 Jul 30;478(14):2889-2898. doi: 10.1042/BCJ20200958. Biochem J. 2021. PMID: 34319404 Free PMC article.
-
Robust dengue virus infection in bat cells and limited innate immune responses coupled with positive serology from bats in IndoMalaya and Australasia.Cell Mol Life Sci. 2020 Apr;77(8):1607-1622. doi: 10.1007/s00018-019-03242-x. Epub 2019 Jul 27. Cell Mol Life Sci. 2020. PMID: 31352533 Free PMC article.
-
A metaanalysis of bat phylogenetics and positive selection based on genomes and transcriptomes from 18 species.Proc Natl Acad Sci U S A. 2019 Jun 4;116(23):11351-11360. doi: 10.1073/pnas.1814995116. Epub 2019 May 21. Proc Natl Acad Sci U S A. 2019. PMID: 31113885 Free PMC article.
References
-
- Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29(3):222–30. - PubMed
-
- Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria central role of complex III. J Biol Chem. 2003;278(38):36027–31. - PubMed
-
- Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59(3):527–605. - PubMed
-
- Kohen R, Nyska A. Invited review: Oxidation of biological systems: oxidative stress phenomena, antioxidants, redox reactions, and methods for their quantification. Toxicol Pathol. 2002;30(6):620–50. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

