A model for genesis of transcription systems
- PMID: 26735411
- PMCID: PMC4802759
- DOI: 10.1080/21541264.2015.1128518
A model for genesis of transcription systems
Abstract
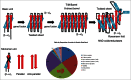
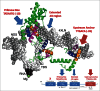
Repeating sequences generated from RNA gene fusions/ligations dominate ancient life, indicating central importance of building structural complexity in evolving biological systems. A simple and coherent story of life on earth is told from tracking repeating motifs that generate α/β proteins, 2-double-Ψ-β-barrel (DPBB) type RNA polymerases (RNAPs), general transcription factors (GTFs), and promoters. A general rule that emerges is that biological complexity that arises through generation of repeats is often bounded by solubility and closure (i.e., to form a pseudo-dimer or a barrel). Because the first DNA genomes were replicated by DNA template-dependent RNA synthesis followed by RNA template-dependent DNA synthesis via reverse transcriptase, the first DNA replication origins were initially 2-DPBB type RNAP promoters. A simplifying model for evolution of promoters/replication origins via repetition of core promoter elements is proposed. The model can explain why Pribnow boxes in bacterial transcription (i.e., (-12)TATAATG(-6)) so closely resemble TATA boxes (i.e., (-31)TATAAAAG(-24)) in archaeal/eukaryotic transcription. The evolution of anchor DNA sequences in bacterial (i.e., (-35)TTGACA(-30)) and archaeal (BRE(up); BRE for TFB recognition element) promoters is potentially explained. The evolution of BRE(down) elements of archaeal promoters is potentially explained.
Keywords: LECA (the last eukaryotic common ancestor); LUCA (the last universal common cellular ancestor); RIFT barrels; RNA polymerase; Rossmann folds; TATA-binding protein (TBP); TIM barrels; cradle-loop barrel metafold; double-Ψ−β-barrels; general transcription factors; replication; the carboxy terminal domain (CTD) of RNA polymerase II; transcription; transcription factor B (TFB); α/β protein folds; σ factors.
Figures





References
-
- Soding J, Lupas AN. More than the sum of their parts: on the evolution of proteins from peptides. BioEssays 2003; 25:837-46; PMID:12938173; http://dx.doi.org/10.1002/bies.10321 - DOI - PubMed
-
- Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol 2003; 3:1; PMID:12553882; http://dx.doi.org/10.1186/1472-6807-3-1 - DOI - PMC - PubMed
-
- Iyer LM, Aravind L. Insights from the architecture of the bacterial transcription apparatus. J Struct Biol 2012; 179:299-319; PMID:22210308; http://dx.doi.org/10.1016/j.jsb.2011.12.013 - DOI - PMC - PubMed
-
- Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J Mol Biol 2010; 395:671-85; PMID:19895820; http://dx.doi.org/10.1016/j.jmb.2009.10.062 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
