Structural Based Analyses of the JC Virus T-Antigen F258L Mutant Provides Evidence for DNA Dependent Conformational Changes in the C-Termini of Polyomavirus Origin Binding Domains
- PMID: 26735515
- PMCID: PMC4703215
- DOI: 10.1371/journal.ppat.1005362
Structural Based Analyses of the JC Virus T-Antigen F258L Mutant Provides Evidence for DNA Dependent Conformational Changes in the C-Termini of Polyomavirus Origin Binding Domains
Erratum in
-
Correction: Structural Based Analyses of the JC Virus T-Antigen F258L Mutant Provides Evidence for DNA Dependent Conformational Changes in the C-Termini of Polyomavirus Origin Binding Domains.PLoS Pathog. 2016 Feb 29;12(2):e1005482. doi: 10.1371/journal.ppat.1005482. eCollection 2016 Feb. PLoS Pathog. 2016. PMID: 26928924 Free PMC article. No abstract available.
Abstract
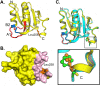
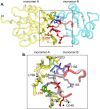
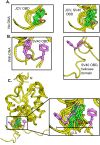
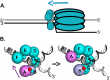
The replication of human polyomavirus JCV, which causes Progressive Multifocal Leukoencephalopathy, is initiated by the virally encoded T-antigen (T-ag). The structure of the JC virus T-ag origin-binding domain (OBD) was recently solved by X-ray crystallography. This structure revealed that the OBD contains a C-terminal pocket, and that residues from the multifunctional A1 and B2 motifs situated on a neighboring OBD molecule dock into the pocket. Related studies established that a mutation in a pocket residue (F258L) rendered JCV T-ag unable to support JCV DNA replication. To establish why this mutation inactivated JCV T-ag, we have solved the structure of the F258L JCV T-ag OBD mutant. Based on this structure, it is concluded that the structural consequences of the F258L mutation are limited to the pocket region. Further analyses, utilizing the available polyomavirus OBD structures, indicate that the F258 region is highly dynamic and that the relative positions of F258 are governed by DNA binding. The possible functional consequences of the DNA dependent rearrangements, including promotion of OBD cycling at the replication fork, are discussed.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Ehlers B, Wieland U. The novel human polyomaviruses HPyV6, 7, 9 and beyond. APMIS. 2013; 10.1111:1–13. - PubMed
-
- Scuda N, Madinda NF, Akoua-Koffi C, Adjogous EV, Wevers D, Hoffman J, et al. Novel Polyomaviruses of Nonhuman Primates: Genetic and Serological Predictors for the Existence of Multiple Unknown Polyomaviruses within the Human Population. PLoS Pathogens. 2013;9:e1003429 10.1371/journal.ppat.1003429 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

