RUNX1 and FOXP3 interplay regulates expression of breast cancer related genes
- PMID: 26735887
- PMCID: PMC4872732
- DOI: 10.18632/oncotarget.6771
RUNX1 and FOXP3 interplay regulates expression of breast cancer related genes
Abstract
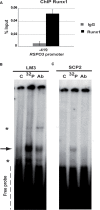
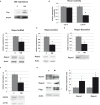
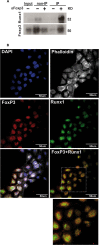
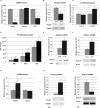
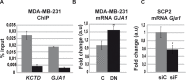
Runx1 participation in epithelial mammary cells is still under review. Emerging data indicates that Runx1 could be relevant for breast tumor promotion. However, to date no studies have specifically evaluated the functional contribution of Runx1 to control gene expression in mammary epithelial tumor cells. It has been described that Runx1 activity is defined by protein context interaction. Interestingly, Foxp3 is a breast tumor suppressor gene. Here we show that endogenous Runx1 and Foxp3 physically interact in normal mammary cells and this interaction blocks Runx1 transcriptional activity. Furthermore we demonstrate that Runx1 is able to bind to R-spondin 3 (RSPO3) and Gap Junction protein Alpha 1 (GJA1) promoters. This binding upregulates Rspo3 oncogene expression and downregulates GJA1 tumor suppressor gene expression in a Foxp3-dependent manner. Moreover, reduced Runx1 transcriptional activity decreases tumor cell migration properties. Collectively, these data provide evidence of a new mechanism for breast tumor gene expression regulation, in which Runx1 and Foxp3 physically interact to control mammary epithelial cell gene expression fate. Our work suggests for the first time that Runx1 could be involved in breast tumor progression depending on Foxp3 availability.
Keywords: Foxp3; GJA1; Rspo3; Runx1; gene expression regulation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Downregulation of Ezh2 methyltransferase by FOXP3: new insight of FOXP3 into chromatin remodeling?Biochim Biophys Acta. 2013 Oct;1833(10):2190-200. doi: 10.1016/j.bbamcr.2013.05.014. Epub 2013 May 17. Biochim Biophys Acta. 2013. PMID: 23688634
-
miR-215 promotes malignant progression of gastric cancer by targeting RUNX1.Oncotarget. 2016 Jan 26;7(4):4817-28. doi: 10.18632/oncotarget.6736. Oncotarget. 2016. PMID: 26716895 Free PMC article.
-
Runx1 stabilizes the mammary epithelial cell phenotype and prevents epithelial to mesenchymal transition.Oncotarget. 2017 Mar 14;8(11):17610-17627. doi: 10.18632/oncotarget.15381. Oncotarget. 2017. PMID: 28407681 Free PMC article.
-
The enigmatic role of RUNX1 in female-related cancers - current knowledge & future perspectives.FEBS J. 2017 Aug;284(15):2345-2362. doi: 10.1111/febs.14059. Epub 2017 Apr 10. FEBS J. 2017. PMID: 28304148 Review.
-
RUNX1 as a Novel Molecular Target for Breast Cancer.Clin Breast Cancer. 2022 Aug;22(6):499-506. doi: 10.1016/j.clbc.2022.04.006. Epub 2022 Apr 26. Clin Breast Cancer. 2022. PMID: 35599145 Review.
Cited by
-
Molecular networks of FOXP family: dual biologic functions, interplay with other molecules and clinical implications in cancer progression.Mol Cancer. 2019 Dec 9;18(1):180. doi: 10.1186/s12943-019-1110-3. Mol Cancer. 2019. PMID: 31815635 Free PMC article. Review.
-
Tumor-infiltrating lymphocytes predict prognosis of breast cancer patients treated with anti-Her-2 therapy.Oncotarget. 2017 Jan 17;8(3):5219-5232. doi: 10.18632/oncotarget.14124. Oncotarget. 2017. PMID: 28029650 Free PMC article.
-
Suppression of Breast Cancer Stem Cells and Tumor Growth by the RUNX1 Transcription Factor.Mol Cancer Res. 2018 Dec;16(12):1952-1964. doi: 10.1158/1541-7786.MCR-18-0135. Epub 2018 Aug 6. Mol Cancer Res. 2018. PMID: 30082484 Free PMC article.
-
Effect of RUNX1/FOXP3 axis on apoptosis of T and B lymphocytes and immunosuppression in sepsis.Open Med (Wars). 2023 Aug 24;18(1):20230728. doi: 10.1515/med-2023-0728. eCollection 2023. Open Med (Wars). 2023. PMID: 37636994 Free PMC article.
-
Direct interaction of Ikaros and Foxp1 modulates expression of the G protein-coupled receptor G2A in B-lymphocytes and acute lymphoblastic leukemia.Oncotarget. 2016 Oct 4;7(40):65923-65936. doi: 10.18632/oncotarget.11688. Oncotarget. 2016. PMID: 27588474 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

