Species difference in ANP32A underlies influenza A virus polymerase host restriction
- PMID: 26738596
- PMCID: PMC4710677
- DOI: 10.1038/nature16474
Species difference in ANP32A underlies influenza A virus polymerase host restriction
Abstract
Influenza pandemics occur unpredictably when zoonotic influenza viruses with novel antigenicity acquire the ability to transmit amongst humans. Host range breaches are limited by incompatibilities between avian virus components and the human host. Barriers include receptor preference, virion stability and poor activity of the avian virus RNA-dependent RNA polymerase in human cells. Mutants of the heterotrimeric viral polymerase components, particularly PB2 protein, are selected during mammalian adaptation, but their mode of action is unknown. We show that a species-specific difference in host protein ANP32A accounts for the suboptimal function of avian virus polymerase in mammalian cells. Avian ANP32A possesses an additional 33 amino acids between the leucine-rich repeats and carboxy-terminal low-complexity acidic region domains. In mammalian cells, avian ANP32A rescued the suboptimal function of avian virus polymerase to levels similar to mammalian-adapted polymerase. Deletion of the avian-specific sequence from chicken ANP32A abrogated this activity, whereas its insertion into human ANP32A, or closely related ANP32B, supported avian virus polymerase function. Substitutions, such as PB2(E627K), were rapidly selected upon infection of humans with avian H5N1 or H7N9 influenza viruses, adapting the viral polymerase for the shorter mammalian ANP32A. Thus ANP32A represents an essential host partner co-opted to support influenza virus replication and is a candidate host target for novel antivirals.
Figures


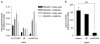
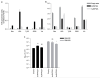



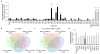

Comment in
-
Virology: Host protein clips bird flu's wings in mammals.Nature. 2016 Jan 7;529(7584):30-1. doi: 10.1038/529030a. Nature. 2016. PMID: 26738587 No abstract available.
-
The Avian Influenza Virus Polymerase Brings ANP32A Home to Roost.Cell Host Microbe. 2016 Feb 10;19(2):137-8. doi: 10.1016/j.chom.2016.01.013. Cell Host Microbe. 2016. PMID: 26867171
References
-
- Cauldwell AV, Long JS, Moncorgé O, Barclay WS. Viral determinants of influenza A virus host range. J. Gen. Virol. 2014;95:1193–210. - PubMed
-
- Almond JW. A single gene determines the host range of influenza virus. Nature. 1977;270:617–8. - PubMed
-
- Naffakh N, Tomoiu A, Rameix-Welti M-A, van der Werf S. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu. Rev. Microbiol. 2008;62:403–24. - PubMed
Additional references for methods
-
- Elleman CJ, Barclay WS. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology. 2004;321:144–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 087039/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- 087039/WT_/Wellcome Trust/United Kingdom
- BB/K002465/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0600006/MRC_/Medical Research Council/United Kingdom
- BBS/E/I/00001708/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

