Activation of RNase L by Murine Coronavirus in Myeloid Cells Is Dependent on Basal Oas Gene Expression and Independent of Virus-Induced Interferon
- PMID: 26739051
- PMCID: PMC4810646
- DOI: 10.1128/JVI.03036-15
Activation of RNase L by Murine Coronavirus in Myeloid Cells Is Dependent on Basal Oas Gene Expression and Independent of Virus-Induced Interferon
Abstract
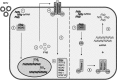
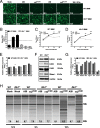
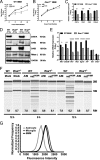
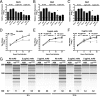
The oligoadenylate synthetase (OAS)-RNase L pathway is a potent interferon (IFN)-induced antiviral activity. Upon sensing double-stranded RNA, OAS produces 2',5'-oligoadenylates (2-5A), which activate RNase L. Murine coronavirus (mouse hepatitis virus [MHV]) nonstructural protein 2 (ns2) is a 2',5'-phosphodiesterase (PDE) that cleaves 2-5A, thereby antagonizing RNase L activation. PDE activity is required for robust replication in myeloid cells, as a mutant of MHV (ns2(H126R)) encoding an inactive PDE fails to antagonize RNase L activation and replicates poorly in bone marrow-derived macrophages (BMM), while ns2(H126R) replicates to high titer in several types of nonmyeloid cells, as well as in IFN receptor-deficient (Ifnar1(-/-)) BMM. We reported previously that myeloid cells express significantly higher basal levels of OAS transcripts than nonmyeloid cells. Here, we investigated the contributions of Oas gene expression, basal IFN signaling, and virus-induced IFN to RNase L activation. Infection with ns2(H126R) activated RNase L in Ifih1(-/-) BMM to a similar extent as in wild-type (WT) BMM, despite the lack of IFN induction in the absence of MDA5 expression. However, ns2(H126R) failed to induce RNase L activation in BMM treated with IFNAR1-blocking antibody, as well as in Ifnar1(-/-) BMM, both expressing low basal levels of Oas genes. Thus, activation of RNase L does not require virus-induced IFN but rather correlates with adequate levels of basal Oas gene expression, maintained by basal IFN signaling. Finally, overexpression of RNase L is not sufficient to compensate for inadequate basal OAS levels.
Importance: The oligoadenylate synthetase (OAS)-RNase L pathway is a potent antiviral activity. Activation of RNase L during murine coronavirus (mouse hepatitis virus [MHV]) infection of myeloid cells correlates with high basal Oas gene expression and is independent of virus-induced interferon secretion. Thus, our data suggest that cells with high basal Oas gene expression levels can activate RNase L and thereby inhibit virus replication early in infection upon exposure to viral double-stranded RNA (dsRNA) before the induction of interferon and prior to transcription of interferon-stimulated antiviral genes. These findings challenge the notion that activation of the OAS-RNase L pathway requires virus to induce type I IFN, which in turn upregulates OAS gene expression, as well as to provide dsRNA to activate OAS. Our data further suggest that myeloid cells may serve as sentinels to restrict viral replication, thus protecting other cell types from infection.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- R01-NS054695/NS/NINDS NIH HHS/United States
- T32 AI007324/AI/NIAID NIH HHS/United States
- T32-NS-007180/NS/NINDS NIH HHS/United States
- T32 NS007180/NS/NINDS NIH HHS/United States
- R01 AI104887/AI/NIAID NIH HHS/United States
- R01-NS081008/NS/NINDS NIH HHS/United States
- T32-AI007324/AI/NIAID NIH HHS/United States
- R01 NS081008/NS/NINDS NIH HHS/United States
- R01-CA044059/CA/NCI NIH HHS/United States
- R01 CA044059/CA/NCI NIH HHS/United States
- R01 NS054695/NS/NINDS NIH HHS/United States
- R01-AI104887/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

