Functional Interplay of Two Paralogs Encoding SWI/SNF Chromatin-Remodeling Accessory Subunits During Caenorhabditis elegans Development
- PMID: 26739451
- PMCID: PMC4788132
- DOI: 10.1534/genetics.115.183533
Functional Interplay of Two Paralogs Encoding SWI/SNF Chromatin-Remodeling Accessory Subunits During Caenorhabditis elegans Development
Abstract
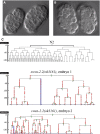
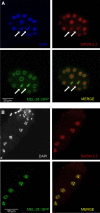
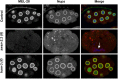
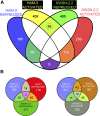
SWI/SNF ATP-dependent chromatin-remodeling complexes have been related to several cellular processes such as transcription, regulation of chromosomal stability, and DNA repair. The Caenorhabditis elegans gene ham-3 (also known as swsn-2.1) and its paralog swsn-2.2 encode accessory subunits of SWI/SNF complexes. Using RNA interference (RNAi) assays and diverse alleles we investigated whether ham-3 and swsn-2.2 have different functions during C. elegans development since they encode proteins that are probably mutually exclusive in a given SWI/SNF complex. We found that ham-3 and swsn-2.2 display similar functions in vulva specification, germline development, and intestinal cell proliferation, but have distinct roles in embryonic development. Accordingly, we detected functional redundancy in some developmental processes and demonstrated by RNA sequencing of RNAi-treated L4 animals that ham-3 and swsn-2.2 regulate the expression of a common subset of genes but also have specific targets. Cell lineage analyses in the embryo revealed hyper-proliferation of intestinal cells in ham-3 null mutants whereas swsn-2.2 is required for proper cell divisions. Using a proteomic approach, we identified SWSN-2.2-interacting proteins needed for early cell divisions, such as SAO-1 and ATX-2, and also nuclear envelope proteins such as MEL-28. swsn-2.2 mutants phenocopy mel-28 loss-of-function, and we observed that SWSN-2.2 and MEL-28 colocalize in mitotic and meiotic chromosomes. Moreover, we demonstrated that SWSN-2.2 is required for correct chromosome segregation and nuclear reassembly after mitosis including recruitment of MEL-28 to the nuclear periphery.
Keywords: Caenorhabditis elegans; SWI/SNF; chromatin; development; nuclear envelope.
Copyright © 2016 by the Genetics Society of America.
Figures

References
-
- Askjaer P., Galy V., Meister P., 2014. Modern tools to study nuclear pore complexes and nucleocytoplasmic transport in Caenorhabditis elegans. Methods Cell Biol. 122: 277–310. - PubMed
-
- Berdasco M., Esteller M., 2013. Genetic syndromes caused by mutations in epigenetic genes. Hum. Genet. 132: 359–383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

