A Pore Idea: the ion conduction pathway of TMEM16/ANO proteins is composed partly of lipid
- PMID: 26739711
- PMCID: PMC4751199
- DOI: 10.1007/s00424-015-1777-2
A Pore Idea: the ion conduction pathway of TMEM16/ANO proteins is composed partly of lipid
Abstract
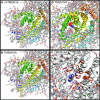
Since their first descriptions, ion channels have been conceived as proteinaceous conduits that facilitate the passage of ionic cargo between segregated environments. This concept is reinforced by crystallographic structures of cation channels depicting ion conductance pathways completely lined by protein. Although lipids are sometimes present in fenestrations near the pore or may be involved in channel gating, there is little or no evidence that lipids inhabit the ion conduction pathway. Indeed, the presence of lipid acyl chains in the conductance pathway would curse the design of the channel's aqueous pore. Here, we make a speculative proposal that anion channels in the TMEM16/ANO superfamily have ion conductance pathways composed partly of lipids. Our reasoning is based on the idea that TMEM16 ion channels evolved from a kind of lipid transporter that scrambles lipids between leaflets of the membrane bilayer and the modeled structural similarity between TMEM16 lipid scramblases and TMEM16 anion channels. This novel view of the TMEM16 pore offers explanation for the biophysical and pharmacological oddness of TMEM16A. We build upon the recent X-ray structure of nhTMEM16 and develop models of both TMEM16 ion channels and lipid scramblases to bolster our proposal. It is our hope that this model of the TMEM16 pore will foster innovative investigation into TMEM16 function.
Keywords: Anoctamin; Calcium; Chloride channel; Phospholipid scrambling; Protein-lipid interactions; TMEM16.
Figures

Similar articles
-
Regulation of TMEM16A/ANO1 and TMEM16F/ANO6 ion currents and phospholipid scrambling by Ca2+ and plasma membrane lipid.J Physiol. 2018 Jan 15;596(2):217-229. doi: 10.1113/JP275175. Epub 2017 Dec 18. J Physiol. 2018. PMID: 29134661 Free PMC article.
-
X-ray structure of a calcium-activated TMEM16 lipid scramblase.Nature. 2014 Dec 11;516(7530):207-12. doi: 10.1038/nature13984. Epub 2014 Nov 12. Nature. 2014. PMID: 25383531
-
Structure and Function of Calcium-Activated Chloride Channels and Phospholipid Scramblases in the TMEM16 Family.Handb Exp Pharmacol. 2024;283:153-180. doi: 10.1007/164_2022_595. Handb Exp Pharmacol. 2024. PMID: 35792944 Review.
-
TMEM16 proteins: unknown structure and confusing functions.J Mol Biol. 2015 Jan 16;427(1):94-105. doi: 10.1016/j.jmb.2014.09.028. Epub 2014 Oct 17. J Mol Biol. 2015. PMID: 25451786 Free PMC article. Review.
-
Putative pore-loops of TMEM16/anoctamin channels affect channel density in cell membranes.J Physiol. 2013 Jul 15;591(14):3487-505. doi: 10.1113/jphysiol.2013.251660. Epub 2013 Apr 22. J Physiol. 2013. PMID: 23613533 Free PMC article.
Cited by
-
Gating mechanism of the extracellular entry to the lipid pathway in a TMEM16 scramblase.Nat Commun. 2018 Aug 14;9(1):3251. doi: 10.1038/s41467-018-05724-1. Nat Commun. 2018. PMID: 30108217 Free PMC article.
-
Known structures and unknown mechanisms of TMEM16 scramblases and channels.J Gen Physiol. 2018 Jul 2;150(7):933-947. doi: 10.1085/jgp.201711957. Epub 2018 Jun 18. J Gen Physiol. 2018. PMID: 29915161 Free PMC article. Review.
-
Anoctamins/TMEM16 Proteins: Chloride Channels Flirting with Lipids and Extracellular Vesicles.Annu Rev Physiol. 2017 Feb 10;79:119-143. doi: 10.1146/annurev-physiol-022516-034031. Epub 2016 Nov 16. Annu Rev Physiol. 2017. PMID: 27860832 Free PMC article. Review.
-
Wide-ranging cellular functions of ion channels and lipid scramblases in the structurally related TMC, TMEM16 and TMEM63 families.Nat Struct Mol Biol. 2025 Feb;32(2):222-236. doi: 10.1038/s41594-024-01444-x. Epub 2024 Dec 23. Nat Struct Mol Biol. 2025. PMID: 39715905 Review.
-
Molecular mechanisms of ion conduction and ion selectivity in TMEM16 lipid scramblases.Nat Commun. 2021 May 14;12(1):2826. doi: 10.1038/s41467-021-22724-w. Nat Commun. 2021. PMID: 33990555 Free PMC article.
References
-
- Andersen T, Haugen HK, Hotop H (1999) Binding energies in atomic negative ions: III. J Phys Chem Ref Data 28:1511–1533, doi:http://dx.doi.org/10.1063/1.556047 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

