Combined deletion of Xrcc4 and Trp53 in mouse germinal center B cells leads to novel B cell lymphomas with clonal heterogeneity
- PMID: 26740101
- PMCID: PMC4704435
- DOI: 10.1186/s13045-015-0230-5
Combined deletion of Xrcc4 and Trp53 in mouse germinal center B cells leads to novel B cell lymphomas with clonal heterogeneity
Abstract
Background: Activated B lymphocytes harbor programmed DNA double-strand breaks (DSBs) initiated by activation-induced deaminase (AID) and repaired by non-homologous end-joining (NHEJ). While it has been proposed that these DSBs during secondary antibody gene diversification are the primary source of chromosomal translocations in germinal center (GC)-derived B cell lymphomas, this point has not been directly addressed due to the lack of proper mouse models.
Methods: In the current study, we establish a unique mouse model by specifically deleting a NHEJ gene, Xrcc4, and a cell cycle checkpoint gene, Trp53, in GC B cells, which results in the spontaneous development of B cell lymphomas that possess features of GC B cells.
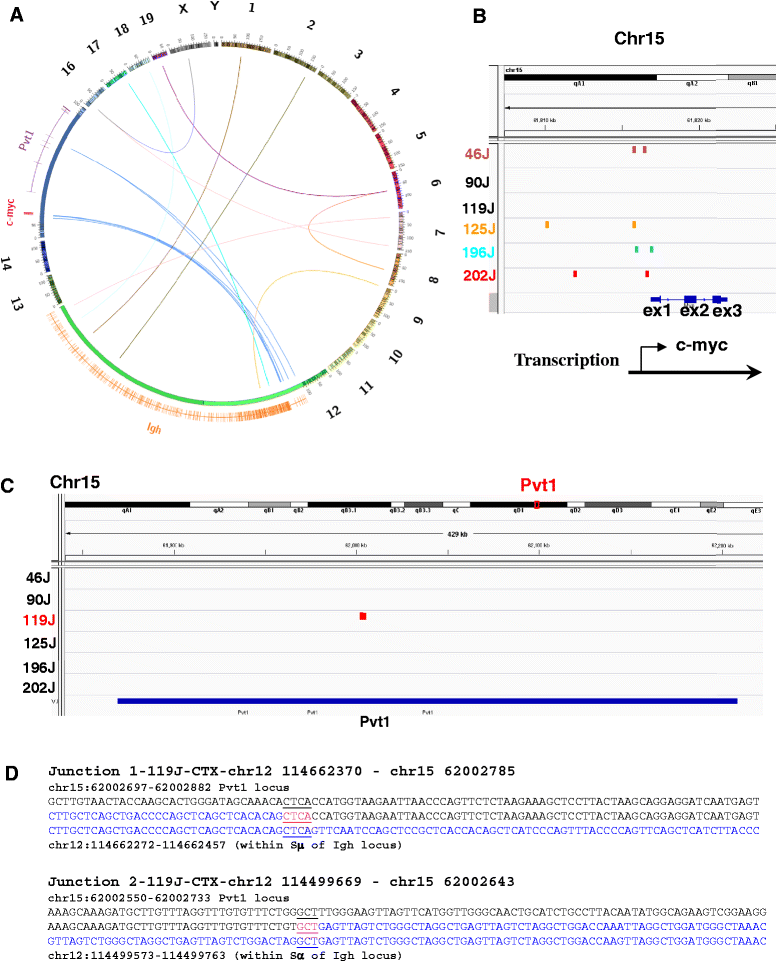
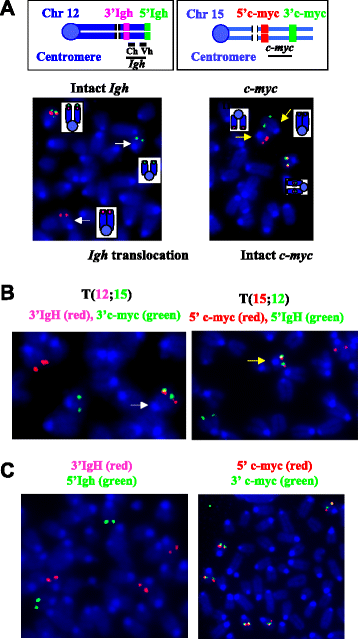
Results: We show that these NHEJ deficient lymphomas harbor translocations frequently targeting immunoglobulin (Ig) loci. Furthermore, we found that Ig translocations were associated with distinct mechanisms, probably caused by AID- or RAG-induced DSBs. Intriguingly, the AID-associated Ig loci translocations target either c-myc or Pvt-1 locus whereas the partners of RAG-associated Ig translocations scattered randomly in the genome. Lastly, these NHEJ deficient lymphomas harbor complicated genomes including segmental translocations and exhibit a high level of ongoing DNA damage and clonal heterogeneity.
Conclusions: We propose that combined NHEJ and p53 defects may serve as an underlying mechanism for a high level of genomic complexity and clonal heterogeneity in cancers.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

