A Comprehensive Genomic Analysis Reveals the Genetic Landscape of Mitochondrial Respiratory Chain Complex Deficiencies
- PMID: 26741492
- PMCID: PMC4704781
- DOI: 10.1371/journal.pgen.1005679
A Comprehensive Genomic Analysis Reveals the Genetic Landscape of Mitochondrial Respiratory Chain Complex Deficiencies
Abstract
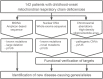
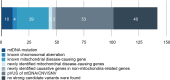
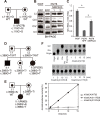
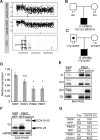
Mitochondrial disorders have the highest incidence among congenital metabolic disorders characterized by biochemical respiratory chain complex deficiencies. It occurs at a rate of 1 in 5,000 births, and has phenotypic and genetic heterogeneity. Mutations in about 1,500 nuclear encoded mitochondrial proteins may cause mitochondrial dysfunction of energy production and mitochondrial disorders. More than 250 genes that cause mitochondrial disorders have been reported to date. However exact genetic diagnosis for patients still remained largely unknown. To reveal this heterogeneity, we performed comprehensive genomic analyses for 142 patients with childhood-onset mitochondrial respiratory chain complex deficiencies. The approach includes whole mtDNA and exome analyses using high-throughput sequencing, and chromosomal aberration analyses using high-density oligonucleotide arrays. We identified 37 novel mutations in known mitochondrial disease genes and 3 mitochondria-related genes (MRPS23, QRSL1, and PNPLA4) as novel causative genes. We also identified 2 genes known to cause monogenic diseases (MECP2 and TNNI3) and 3 chromosomal aberrations (6q24.3-q25.1, 17p12, and 22q11.21) as causes in this cohort. Our approaches enhance the ability to identify pathogenic gene mutations in patients with biochemically defined mitochondrial respiratory chain complex deficiencies in clinical settings. They also underscore clinical and genetic heterogeneity and will improve patient care of this complex disorder.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

