PUMA promotes apoptosis of hematopoietic progenitors driving leukemic progression in a mouse model of myelodysplasia
- PMID: 26742432
- PMCID: PMC4987724
- DOI: 10.1038/cdd.2015.159
PUMA promotes apoptosis of hematopoietic progenitors driving leukemic progression in a mouse model of myelodysplasia
Abstract
Myelodysplastic syndrome (MDS) is characterized by ineffective hematopoiesis with resultant cytopenias. Increased apoptosis and aberrantly functioning progenitors are thought to contribute to this phenotype. As is the case for other malignancies, overcoming apoptosis is believed to be important in progression toward acute myeloid leukemia (AML). Using the NUP98-HOXD13 (NHD13) transgenic mouse model of MDS, we previously reported that overexpression of the anti-apoptotic protein BCL2, blocked apoptosis and improved cytopenias, paradoxically, delaying leukemic progression. To further understand this surprising result, we examined the role of p53 and its pro-apoptotic effectors, PUMA and NOXA in NHD13 mice. The absence of p53 or PUMA but not NOXA reduced apoptosis and expanded the numbers of MDS-repopulating cells. Despite a similar effect on apoptosis and cell numbers, the absence of p53 and PUMA had diametrically opposed effects on progression to AML: absence of p53 accelerated leukemic progression, while absence of PUMA significantly delayed progression. This may be explained in part by differences in cellular responses to DNA damage. The absence of p53 led to higher levels of γ-H2AX (indicative of persistent DNA lesions) while PUMA-deficient NHD13 progenitors resolved DNA lesions in a manner comparable to wild-type cells. These results suggest that targeting PUMA may improve the cytopenias of MDS without a detrimental effect on leukemic progression thus warranting further investigation.
Figures



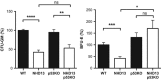
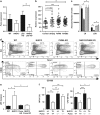
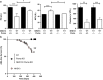
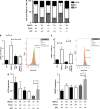
References
-
- Woll PS, Kjällquist U, Chowdhury O, Doolittle H, Wedge DC, Thongjuea S et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell 2014; 25: 794–808. - PubMed
-
- Raza A, Galili N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat Rev Cancer 2012; 12: 849–859. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

