De Novo Transcriptome Analysis of Cucumis melo L. var. makuwa
- PMID: 26743902
- PMCID: PMC4757802
- DOI: 10.14348/molcells.2016.2264
De Novo Transcriptome Analysis of Cucumis melo L. var. makuwa
Abstract
Oriental melon (Cucumis melo L. var. makuwa) is one of six subspecies of melon and is cultivated widely in East Asia, including China, Japan, and Korea. Although oriental melon is economically valuable in Asia and is genetically distinct from other subspecies, few reports of genome-scale research on oriental melon have been published. We generated 30.5 and 36.8 Gb of raw RNA sequence data from the female and male flowers, leaves, roots, and fruit of two oriental melon varieties, Korean landrace (KM) and Breeding line of NongWoo Bio Co. (NW), respectively. From the raw reads, 64,998 transcripts from KM and 100,234 transcripts from NW were de novo assembled. The assembled transcripts were used to identify molecular markers (e.g., single-nucleotide polymorphisms and simple sequence repeats), detect tissue-specific expressed genes, and construct a genetic linkage map. In total, 234 single-nucleotide polymorphisms and 25 simple sequence repeats were screened from 7,871 and 8,052 candidates, respectively, between the KM and NW varieties and used for construction of a genetic map with 94 F2 population specimens. The genetic linkage map consisted of 12 linkage groups, and 248 markers were assigned. These transcriptome and molecular marker data provide information useful for molecular breeding of oriental melon and further comparative studies of the Cucurbitaceae family.
Keywords: Korean melon; genetic linkage map; simple sequence repeat; single-nucleotide polymorphism; transcriptome analysis.
Figures
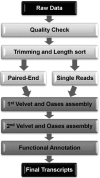
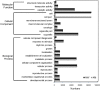
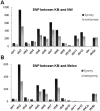

References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Blanca J.M., Cañizares J., Ziarsolo P., Esteras C., Mir G., Nuez F., Garcia-Mas J., Picó M.B. Melon transcriptome characterization: simple sequence repeats and single nucleotide polymorphisms discovery for high throughput genotyping across the species. Plant Genome. 2011;4:118–131.
-
- Brisson N., Paszkowski J., Penswick J.R., Gronenborn B., Potrykus I., Hohn T. Expression of a bacterial gene in plants by using a viral vector. Nature. 1984;310:511–514.
-
- Cornejo M.J., Luth D., Blankenship K.M., Anderson O.D., Blechl A.E. Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol. Biol. 1993;23:567–581. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

