The Floating (Pathogenicity) Island: A Genomic Dessert
- PMID: 26744223
- PMCID: PMC4733582
- DOI: 10.1016/j.tig.2015.11.005
The Floating (Pathogenicity) Island: A Genomic Dessert
Abstract
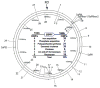
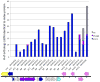
Among the prokaryotic genomic islands (GIs) involved in horizontal gene transfer (HGT) are the classical pathogenicity islands, including the integrative and conjugative elements (ICEs), the gene-transfer agents (GTAs), and the staphylococcal pathogenicity islands (SaPIs), the primary focus of this review. While the ICEs and GTAs mediate HGT autonomously, the SaPIs are dependent on specific phages. The ICEs transfer primarily their own DNA, the GTAs exclusively transfer unlinked host DNA, and the SaPIs combine the capabilities of both. Thus the SaPIs derive their importance from the genes they carry (their genetic cargo) and the genes they move. They act not only as versatile high-frequency mobilizers but also as mediators of phage interference and consequently are major benefactors of their host bacteria.
Keywords: Staphylococcus; horizontal gene transfer; mobile genetic element; pathogenicity island; phage interference; transduction.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


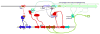



Similar articles
-
Staphylococcal pathogenicity islands-movers and shakers in the genomic firmament.Curr Opin Microbiol. 2017 Aug;38:197-204. doi: 10.1016/j.mib.2017.08.001. Epub 2017 Nov 1. Curr Opin Microbiol. 2017. PMID: 29100762 Free PMC article. Review.
-
Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism.Proc Natl Acad Sci U S A. 2012 Oct 2;109(40):16300-5. doi: 10.1073/pnas.1204615109. Epub 2012 Sep 18. Proc Natl Acad Sci U S A. 2012. PMID: 22991467 Free PMC article.
-
The SaPIs: mobile pathogenicity islands of Staphylococcus.Chem Immunol Allergy. 2007;93:42-57. doi: 10.1159/000100857. Chem Immunol Allergy. 2007. PMID: 17369699 Review.
-
Staphylococcal pathogenicity island DNA packaging system involving cos-site packaging and phage-encoded HNH endonucleases.Proc Natl Acad Sci U S A. 2014 Apr 22;111(16):6016-21. doi: 10.1073/pnas.1320538111. Epub 2014 Apr 7. Proc Natl Acad Sci U S A. 2014. PMID: 24711396 Free PMC article.
-
Pathogenicity Islands and Their Role in Staphylococcal Biology.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.gpp3-0062-2019. doi: 10.1128/microbiolspec.GPP3-0062-2019. Microbiol Spectr. 2019. PMID: 31172913 Free PMC article. Review.
Cited by
-
Phage-mediated horizontal gene transfer and its implications for the human gut microbiome.Gastroenterol Rep (Oxf). 2022 Apr 13;10:goac012. doi: 10.1093/gastro/goac012. eCollection 2022. Gastroenterol Rep (Oxf). 2022. PMID: 35425613 Free PMC article.
-
Mobile Genetic Elements Associated with Antimicrobial Resistance.Clin Microbiol Rev. 2018 Aug 1;31(4):e00088-17. doi: 10.1128/CMR.00088-17. Print 2018 Oct. Clin Microbiol Rev. 2018. PMID: 30068738 Free PMC article. Review.
-
Integrated mobile genetic elements in Thaumarchaeota.Environ Microbiol. 2019 Jun;21(6):2056-2078. doi: 10.1111/1462-2920.14564. Epub 2019 Mar 18. Environ Microbiol. 2019. PMID: 30773816 Free PMC article.
-
Competing scaffolding proteins determine capsid size during mobilization of Staphylococcus aureus pathogenicity islands.Elife. 2017 Oct 6;6:e30822. doi: 10.7554/eLife.30822. Elife. 2017. PMID: 28984245 Free PMC article.
-
Genomic analysis of Helicobacter himalayensis sp. nov. isolated from Marmota himalayana.BMC Genomics. 2020 Nov 23;21(1):826. doi: 10.1186/s12864-020-07245-y. BMC Genomics. 2020. PMID: 33228534 Free PMC article.
References
-
- Perna NT, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

