Conserved pharmacological rescue of hereditary spastic paraplegia-related phenotypes across model organisms
- PMID: 26744324
- PMCID: PMC4764191
- DOI: 10.1093/hmg/ddv632
Conserved pharmacological rescue of hereditary spastic paraplegia-related phenotypes across model organisms
Abstract
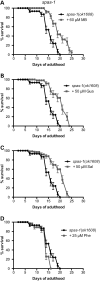
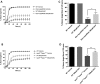
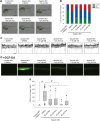
Hereditary spastic paraplegias (HSPs) are a group of neurodegenerative diseases causing progressive gait dysfunction. Over 50 genes have now been associated with HSP. Despite the recent explosion in genetic knowledge, HSP remains without pharmacological treatment. Loss-of-function mutation of the SPAST gene, also known as SPG4, is the most common cause of HSP in patients. SPAST is conserved across animal species and regulates microtubule dynamics. Recent studies have shown that it also modulates endoplasmic reticulum (ER) stress. Here, utilizing null SPAST homologues in C. elegans, Drosophila and zebrafish, we tested FDA-approved compounds known to modulate ER stress in order to ameliorate locomotor phenotypes associated with HSP. We found that locomotor defects found in all of our spastin models could be partially rescued by phenazine, methylene blue, N-acetyl-cysteine, guanabenz and salubrinal. In addition, we show that established biomarkers of ER stress levels correlated with improved locomotor activity upon treatment across model organisms. Our results provide insights into biomarkers and novel therapeutic avenues for HSP.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures






Similar articles
-
Disease-related phenotypes in a Drosophila model of hereditary spastic paraplegia are ameliorated by treatment with vinblastine.J Clin Invest. 2005 Nov;115(11):3026-34. doi: 10.1172/JCI24694. J Clin Invest. 2005. PMID: 16276413 Free PMC article.
-
Hereditary spastic paraplegia SPG4: what is known and not known about the disease.Brain. 2015 Sep;138(Pt 9):2471-84. doi: 10.1093/brain/awv178. Epub 2015 Jun 20. Brain. 2015. PMID: 26094131 Free PMC article. Review.
-
Cul-4 inhibition rescues spastin levels and reduces defects in hereditary spastic paraplegia models.Brain. 2024 Oct 3;147(10):3534-3546. doi: 10.1093/brain/awae095. Brain. 2024. PMID: 38551087 Free PMC article.
-
A patient-derived stem cell model of hereditary spastic paraplegia with SPAST mutations.Dis Model Mech. 2013 Mar;6(2):489-502. doi: 10.1242/dmm.010884. Epub 2012 Dec 20. Dis Model Mech. 2013. PMID: 23264559 Free PMC article.
-
Exon 8-17 deletions of SPAST in a Chinese family with hereditary spastic paraplegia: a case report and literature review.J Neurol Sci. 2015 Oct 15;357(1-2):282-4. doi: 10.1016/j.jns.2015.07.003. Epub 2015 Jul 3. J Neurol Sci. 2015. PMID: 26165777 Review.
Cited by
-
Loss of Fic causes progressive neurodegeneration in a Drosophila model of hereditary spastic paraplegia.Biochim Biophys Acta Mol Basis Dis. 2024 Oct;1870(7):167348. doi: 10.1016/j.bbadis.2024.167348. Epub 2024 Jul 8. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 38986817
-
Targeting Endoplasmic Reticulum Stress as an Effective Treatment for Alcoholic Pancreatitis.Biomedicines. 2022 Jan 5;10(1):108. doi: 10.3390/biomedicines10010108. Biomedicines. 2022. PMID: 35052788 Free PMC article. Review.
-
Dominant negative effect as a novel mechanism of SPAST gene mutation in a large family with hereditary spastic paraplegia.Genes Dis. 2023 Oct 27;11(5):101152. doi: 10.1016/j.gendis.2023.101152. eCollection 2024 Sep. Genes Dis. 2023. PMID: 38882014 Free PMC article. No abstract available.
-
Clinical Trial Designs and Measures in Hereditary Spastic Paraplegias.Front Neurol. 2018 Dec 21;9:1017. doi: 10.3389/fneur.2018.01017. eCollection 2018. Front Neurol. 2018. PMID: 30627115 Free PMC article. Review.
-
Hereditary spastic paraplegia: gain-of-function mechanisms revealed by new transgenic mouse.Hum Mol Genet. 2019 Apr 1;28(7):1136-1152. doi: 10.1093/hmg/ddy419. Hum Mol Genet. 2019. PMID: 30520996 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

