Real-Time Observation of Backtracking by Bacterial RNA Polymerase
- PMID: 26745324
- PMCID: PMC5921838
- DOI: 10.1021/acs.biochem.5b01184
Real-Time Observation of Backtracking by Bacterial RNA Polymerase
Abstract
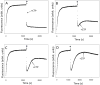
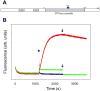
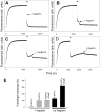
RNA polymerase (RNAP) backtracking is a backward sliding of the enzyme along DNA and RNA. It plays important roles in many essential processes in bacteria and in eukaryotes. We describe here a fluorescence-based approach that allows a real-time observation of bacterial RNAP backtracking. A Cy3 fluorescence probe, when incorporated into a specific site in the nontemplate strand near the site of backtracking, allows RNAP movements to be monitored near the probe because of a robust enhancement of fluorescence caused by protein proximity. Using this approach, we showed that binding of NTP to the active site prior to phosphodiester bond formation inhibited backtracking, consistent with the coupling of NTP binding to translocation. The extent and the kinetics of backtracking did not show a simple correlation with the instability of the DNA-RNA hybrid, indicating a more complex dependence of backtracking on DNA template sequence. Experiments with transcription through an abasic site in DNA template or neutravidin bound to biotinylated template strand base illustrated an important role of backtracking in defining how RNAP reacts to such obstacles in the DNA template. The described approach will be a useful tool in deciphering the mechanism of backtracking and in studying factors that affect the backtracking.
Figures

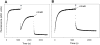



Similar articles
-
Backtracking by single RNA polymerase molecules observed at near-base-pair resolution.Nature. 2003 Dec 11;426(6967):684-7. doi: 10.1038/nature02191. Epub 2003 Nov 23. Nature. 2003. PMID: 14634670 Free PMC article.
-
Maintenance of RNA-DNA hybrid length in bacterial RNA polymerases.J Biol Chem. 2009 May 15;284(20):13497-13504. doi: 10.1074/jbc.M901898200. Epub 2009 Mar 25. J Biol Chem. 2009. PMID: 19321439 Free PMC article.
-
Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking.Cell. 2003 Nov 14;115(4):437-47. doi: 10.1016/s0092-8674(03)00845-6. Cell. 2003. PMID: 14622598
-
Transcription elongation.Transcription. 2017 May 27;8(3):150-161. doi: 10.1080/21541264.2017.1289294. Epub 2017 Feb 8. Transcription. 2017. PMID: 28301288 Free PMC article. Review.
-
Mechanisms of Bacterial Transcription Termination: All Good Things Must End.Annu Rev Biochem. 2016 Jun 2;85:319-47. doi: 10.1146/annurev-biochem-060815-014844. Epub 2016 Mar 17. Annu Rev Biochem. 2016. PMID: 27023849 Review.
Cited by
-
β-CASP proteins removing RNA polymerase from DNA: when a torpedo is needed to shoot a sitting duck.Nucleic Acids Res. 2021 Oct 11;49(18):10221-10234. doi: 10.1093/nar/gkab803. Nucleic Acids Res. 2021. PMID: 34551438 Free PMC article. Review.
-
A new twist on PIFE: photoisomerisation-related fluorescence enhancement.Methods Appl Fluoresc. 2023 Oct 12;12(1):012001. doi: 10.1088/2050-6120/acfb58. Methods Appl Fluoresc. 2023. PMID: 37726007 Free PMC article. Review.
-
Modified nucleoside triphosphates in bacterial research for in vitro and live-cell applications.RSC Chem Biol. 2020 Dec 1;1(5):333-351. doi: 10.1039/d0cb00078g. Epub 2020 Sep 14. RSC Chem Biol. 2020. PMID: 33928252 Free PMC article.
-
A new twist on PIFE: photoisomerisation-related fluorescence enhancement.ArXiv [Preprint]. 2023 Jul 10:arXiv:2302.12455v2. ArXiv. 2023. Update in: Methods Appl Fluoresc. 2023 Oct 12;12(1). doi: 10.1088/2050-6120/acfb58. PMID: 36866225 Free PMC article. Updated. Preprint.
-
DNA template sequence control of bacterial RNA polymerase escape from the promoter.Nucleic Acids Res. 2018 May 18;46(9):4469-4486. doi: 10.1093/nar/gky172. Nucleic Acids Res. 2018. PMID: 29546317 Free PMC article.
References
-
- Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochemical Society transactions. 2006;34:1062–1066. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

