Mechanical Folding and Unfolding of Protein Barnase at the Single-Molecule Level
- PMID: 26745410
- PMCID: PMC4825109
- DOI: 10.1016/j.bpj.2015.11.015
Mechanical Folding and Unfolding of Protein Barnase at the Single-Molecule Level
Abstract
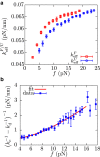
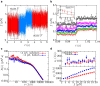
The unfolding and folding of protein barnase has been extensively investigated in bulk conditions under the effect of denaturant and temperature. These experiments provided information about structural and kinetic features of both the native and the unfolded states of the protein, and debates about the possible existence of an intermediate state in the folding pathway have arisen. Here, we investigate the folding/unfolding reaction of protein barnase under the action of mechanical force at the single-molecule level using optical tweezers. We measure unfolding and folding force-dependent kinetic rates from pulling and passive experiments, respectively, and using Kramers-based theories (e.g., Bell-Evans and Dudko-Hummer-Szabo models), we extract the position of the transition state and the height of the kinetic barrier mediating unfolding and folding transitions, finding good agreement with previous bulk measurements. Measurements of the force-dependent kinetic barrier using the continuous effective barrier analysis show that protein barnase verifies the Leffler-Hammond postulate under applied force and allow us to extract its free energy of folding, ΔG0. The estimated value of ΔG0 is in agreement with our predictions obtained using fluctuation relations and previous bulk studies. To address the possible existence of an intermediate state on the folding pathway, we measure the power spectrum of force fluctuations at high temporal resolution (50 kHz) when the protein is either folded or unfolded and, additionally, we study the folding transition-path time at different forces. The finite bandwidth of our experimental setup sets the lifetime of potential intermediate states upon barnase folding/unfolding in the submillisecond timescale.
Copyright © 2016 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Ritort F. Single-molecule experiments in biological physics: methods and applications. J. Phys. Condens. Matter. 2006;18:R531–R583. - PubMed
-
- Dill K.A., Chan H.S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997;4:10–19. - PubMed
-
- Borgia A., Williams P.M., Clarke J. Single-molecule studies of protein folding. Annu. Rev. Biochem. 2008;77:101–125. - PubMed
-
- Zoldák G., Rief M. Force as a single molecule probe of multidimensional protein energy landscapes. Curr. Opin. Struct. Biol. 2013;23:48–57. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

