Structural Analysis on the Pathologic Mutant Glucocorticoid Receptor Ligand-Binding Domains
- PMID: 26745667
- PMCID: PMC4792232
- DOI: 10.1210/me.2015-1177
Structural Analysis on the Pathologic Mutant Glucocorticoid Receptor Ligand-Binding Domains
Abstract
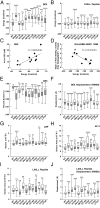
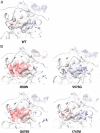
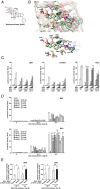
Glucocorticoid receptor (GR) gene mutations may cause familial or sporadic generalized glucocorticoid resistance syndrome. Most of the missense forms distribute in the ligand-binding domain and impair its ligand-binding activity and formation of the activation function (AF)-2 that binds LXXLL motif-containing coactivators. We performed molecular dynamics simulations to ligand-binding domain of pathologic GR mutants to reveal their structural defects. Several calculated parameters including interaction energy for dexamethasone or the LXXLL peptide indicate that destruction of ligand-binding pocket (LBP) is a primary character. Their LBP defects are driven primarily by loss/reduction of the electrostatic interaction formed by R611 and T739 of the receptor to dexamethasone and a subsequent conformational mismatch, which deacylcortivazol resolves with its large phenylpyrazole moiety and efficiently stimulates transcriptional activity of the mutant receptors with LBP defect. Reduced affinity of the LXXLL peptide to AF-2 is caused mainly by disruption of the electrostatic bonds to the noncore leucine residues of this peptide that determine the peptide's specificity to GR, as well as by reduced noncovalent interaction against core leucines and subsequent exposure of the AF-2 surface to solvent. The results reveal molecular defects of pathologic mutant receptors and provide important insights to the actions of wild-type GR.
Figures




References
-
- Chrousos GP. Stress and disorders of the stress system. Nat Rev Endocrinol. 2009;5:374–381. - PubMed
-
- Kino T, Chrousos GP. Glucocorticoid effect on gene expression. In: Steckler T, Kalin NH, Reul JMHM, eds. Handbook on Stress and the Brain. Amsterdam, The Netherlands: Elsevier BV; 2005:295–312.
-
- Chrousos GP, Kino T. Intracellular glucocorticoid signaling: a formerly simple system turns stochastic. Sci STKE. 2005;2005:pe48. - PubMed
-
- Kino T. Glucocorticoid receptor. In: Chrousos GP, ed. Adrenal Disease and Function. South Dartmouth, MA: Endotext; 2000.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

