Arabidopsis ERF1 Mediates Cross-Talk between Ethylene and Auxin Biosynthesis during Primary Root Elongation by Regulating ASA1 Expression
- PMID: 26745809
- PMCID: PMC4706318
- DOI: 10.1371/journal.pgen.1005760
Arabidopsis ERF1 Mediates Cross-Talk between Ethylene and Auxin Biosynthesis during Primary Root Elongation by Regulating ASA1 Expression
Erratum in
-
Correction: Arabidopsis ERF1 Mediates Cross-Talk between Ethylene and Auxin Biosynthesis during Primary Root Elongation by Regulating ASA1 Expression.PLoS Genet. 2016 May 18;12(5):e1006076. doi: 10.1371/journal.pgen.1006076. eCollection 2016 May. PLoS Genet. 2016. PMID: 27191951 Free PMC article.
Abstract
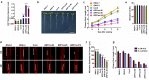
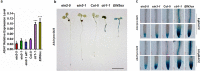
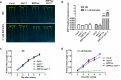
The gaseous phytohormone ethylene participates in the regulation of root growth and development in Arabidopsis. It is known that root growth inhibition by ethylene involves auxin, which is partially mediated by the action of the WEAK ETHYLENE INSENSITIVE2/ANTHRANILATE SYNTHASE α1 (WEI2/ASA1), encoding a rate-limiting enzyme in tryptophan (Trp) biosynthesis, from which auxin is derived. However, the molecular mechanism by which ethylene decreases root growth via ASA1 is not understood. Here we report that the ethylene-responsive AP2 transcription factor, ETHYLENE RESPONSE FACTOR1 (ERF1), plays an important role in primary root elongation of Arabidopsis. Using loss- and gain-of-function transgenic lines as well as biochemical analysis, we demonstrate that ERF1 can directly up-regulate ASA1 by binding to its promoter, leading to auxin accumulation and ethylene-induced inhibition of root growth. This discloses one mechanism linking ethylene signaling and auxin biosynthesis in Arabidopsis roots.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Swarup R, Parry G, Graham N, Allen T, Bennett M (2002) Auxin cross-talk: integration of signalling pathways to control plant development. Plant Mol Biol 49: 411–426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

