Comparative Analysis of Xenorhabdus koppenhoeferi Gene Expression during Symbiotic Persistence in the Host Nematode
- PMID: 26745883
- PMCID: PMC4706420
- DOI: 10.1371/journal.pone.0145739
Comparative Analysis of Xenorhabdus koppenhoeferi Gene Expression during Symbiotic Persistence in the Host Nematode
Abstract
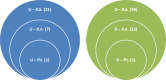
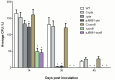
Species of Xenorhabdus and Photorhabdus bacteria form mutualistic associations with Steinernema and Heterorhabditis nematodes, respectively and serve as model systems for studying microbe-animal symbioses. Here, we profiled gene expression of Xenorhabdus koppenhoeferi during their symbiotic persistence in the newly formed infective juveniles of the host nematode Steinernema scarabaei through the selective capture of transcribed sequences (SCOTS). The obtained gene expression profile was then compared with other nematode-bacteria partnerships represented by Steinernema carpocapsae-Xenorhabdus nematophila and Heterorhabditis bacteriophora-Photorhabdus temperata. A total of 29 distinct genes were identified to be up-regulated and 53 were down-regulated in X. koppenhoeferi while in S. scarabaei infective juveniles. Of the identified genes, 8 of the up-regulated and 14 of the down-regulated genes were similarly expressed in X. nematophila during persistence in its host nematode S. carpocapsae. However, only one from each of these up- and down-regulated genes was common to the mutualistic partnership between the bacterium P. temperata and the nematode H. bacteriophora. Interactive network analysis of the shared genes between X. koppenhoeferi and X. nematophila demonstrated that the up-regulated genes were mainly involved in bacterial survival and the down-regulated genes were more related to bacterial virulence and active growth. Disruption of two selected genes pta (coding phosphotransacetylase) and acnB (coding aconitate hydratase) in X. nematophila with shared expression signature with X. koppenhoeferi confirmed that these genes are important for bacterial persistence in the nematode host. The results of our comparative analyses show that the two Xenorhabdus species share a little more than a quarter of the transcriptional mechanisms during persistence in their nematode hosts but these features are quite different from those used by P. temperata bacteria in their nematode host H. bacteriophora.
Conflict of interest statement
Figures


References
-
- Goodrich-Blair H, Clarke DJ (2007) Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol Microbiol 64: 260–268. - PubMed
-
- Stock SP, Goodrich-Blair H (2008) Nematode-bacterium symbioses: Crossing kingdom and disciplinary boundaries. Symbiosis 46: 61–64.
-
- ffrench-Constant R, Waterfield N, Daborn P, Joyce S, Bennett H, Au C, et al. (2003) Photorhabdus: towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiol Rev 26: 433–456. - PubMed
-
- Bird AF, Akhurst RJ (1983) The nature of the intestinal vesicle in nematodes of the family Steinernematidae. Int J Parasitol 13: 599–606.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

