Comparative performance evaluation of automated segmentation methods of hippocampus from magnetic resonance images of temporal lobe epilepsy patients
- PMID: 26745947
- PMCID: PMC4706546
- DOI: 10.1118/1.4938411
Comparative performance evaluation of automated segmentation methods of hippocampus from magnetic resonance images of temporal lobe epilepsy patients
Abstract
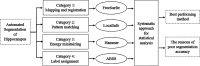
Purpose: Segmentation of the hippocampus from magnetic resonance (MR) images is a key task in the evaluation of mesial temporal lobe epilepsy (mTLE) patients. Several automated algorithms have been proposed although manual segmentation remains the benchmark. Choosing a reliable algorithm is problematic since structural definition pertaining to multiple edges, missing and fuzzy boundaries, and shape changes varies among mTLE subjects. Lack of statistical references and guidance for quantifying the reliability and reproducibility of automated techniques has further detracted from automated approaches. The purpose of this study was to develop a systematic and statistical approach using a large dataset for the evaluation of automated methods and establish a method that would achieve results better approximating those attained by manual tracing in the epileptogenic hippocampus.
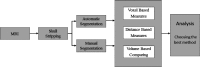
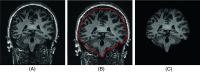
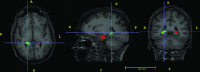
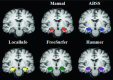
Methods: A template database of 195 (81 males, 114 females; age range 32-67 yr, mean 49.16 yr) MR images of mTLE patients was used in this study. Hippocampal segmentation was accomplished manually and by two well-known tools (FreeSurfer and hammer) and two previously published methods developed at their institution [Automatic brain structure segmentation (ABSS) and LocalInfo]. To establish which method was better performing for mTLE cases, several voxel-based, distance-based, and volume-based performance metrics were considered. Statistical validations of the results using automated techniques were compared with the results of benchmark manual segmentation. Extracted metrics were analyzed to find the method that provided a more similar result relative to the benchmark.
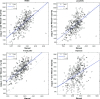
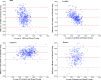
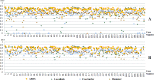
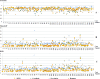
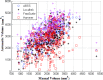
Results: Among the four automated methods, ABSS generated the most accurate results. For this method, the Dice coefficient was 5.13%, 14.10%, and 16.67% higher, Hausdorff was 22.65%, 86.73%, and 69.58% lower, precision was 4.94%, -4.94%, and 12.35% higher, and the root mean square (RMS) was 19.05%, 61.90%, and 65.08% lower than LocalInfo, FreeSurfer, and hammer, respectively. The Bland-Altman similarity analysis revealed a low bias for the ABSS and LocalInfo techniques compared to the others.
Conclusions: The ABSS method for automated hippocampal segmentation outperformed other methods, best approximating what could be achieved by manual tracing. This study also shows that four categories of input data can cause automated segmentation methods to fail. They include incomplete studies, artifact, low signal-to-noise ratio, and inhomogeneity. Different scanner platforms and pulse sequences were considered as means by which to improve reliability of the automated methods. Other modifications were specially devised to enhance a particular method assessed in this study.
Figures


Similar articles
-
Statistical validation of automatic methods for hippocampus segmentation in MR images of epileptic patients.Annu Int Conf IEEE Eng Med Biol Soc. 2014;2014:4707-10. doi: 10.1109/EMBC.2014.6944675. Annu Int Conf IEEE Eng Med Biol Soc. 2014. PMID: 25571043 Free PMC article.
-
Hippocampal volumetry for lateralization of temporal lobe epilepsy: automated versus manual methods.Neuroimage. 2011 Jan;54 Suppl 1:S218-26. doi: 10.1016/j.neuroimage.2010.03.066. Epub 2010 Mar 29. Neuroimage. 2011. PMID: 20353827 Free PMC article.
-
Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates.Neuroimage. 2014 Nov 1;101:494-512. doi: 10.1016/j.neuroimage.2014.04.054. Epub 2014 Apr 29. Neuroimage. 2014. PMID: 24784800
-
Automated methods for hippocampus segmentation: the evolution and a review of the state of the art.Neuroinformatics. 2015 Apr;13(2):133-50. doi: 10.1007/s12021-014-9243-4. Neuroinformatics. 2015. PMID: 26022748 Review.
-
Comparative Approach of MRI-Based Brain Tumor Segmentation and Classification Using Genetic Algorithm.J Digit Imaging. 2018 Aug;31(4):477-489. doi: 10.1007/s10278-018-0050-6. J Digit Imaging. 2018. PMID: 29344753 Free PMC article. Review.
Cited by
-
The bumps under the hippocampus.Hum Brain Mapp. 2018 Jan;39(1):472-490. doi: 10.1002/hbm.23856. Epub 2017 Oct 23. Hum Brain Mapp. 2018. PMID: 29058349 Free PMC article.
-
Multi-atlas label fusion with random local binary pattern features: Application to hippocampus segmentation.Sci Rep. 2019 Nov 14;9(1):16839. doi: 10.1038/s41598-019-53387-9. Sci Rep. 2019. PMID: 31727982 Free PMC article.
-
Hippocampal subfield volumetric changes after radiotherapy for brain metastases.Neurooncol Adv. 2024 Mar 20;6(1):vdae040. doi: 10.1093/noajnl/vdae040. eCollection 2024 Jan-Dec. Neurooncol Adv. 2024. PMID: 38645488 Free PMC article.
-
Clinical validation of automated hippocampal segmentation in temporal lobe epilepsy.Neuroimage Clin. 2018;20:1139-1147. doi: 10.1016/j.nicl.2018.09.032. Epub 2018 Oct 10. Neuroimage Clin. 2018. PMID: 30380521 Free PMC article.
-
Deep Learning for Autism Diagnosis and Facial Analysis in Children.Front Comput Neurosci. 2022 Jan 20;15:789998. doi: 10.3389/fncom.2021.789998. eCollection 2021. Front Comput Neurosci. 2022. Retraction in: Front Comput Neurosci. 2023 May 16;17:1215827. doi: 10.3389/fncom.2023.1215827. PMID: 35126078 Free PMC article. Retracted.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

