IRF4 and IRF8 Act in CD11c+ Cells To Regulate Terminal Differentiation of Lung Tissue Dendritic Cells
- PMID: 26746189
- PMCID: PMC4744567
- DOI: 10.4049/jimmunol.1501870
IRF4 and IRF8 Act in CD11c+ Cells To Regulate Terminal Differentiation of Lung Tissue Dendritic Cells
Abstract
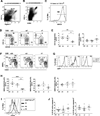
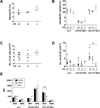
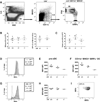
Dendritic cells (DCs) initiate immune responses in barrier tissues including lung and skin. Conventional DC (cDC) subsets, CD11b(-) (cDC1s) or CD11b(+) (cDC2s), arise via distinct networks of transcription factors involving IFN regulatory factor 4 (IRF4) and IRF8, and are specialized for unique functional responses. Using mice in which a conditional Irf4 or Irf8 allele is deleted in CD11c(+) cells, we determined whether IRF4 or IRF8 deficiency beginning in CD11c(+) cDC precursors (pre-cDCs) changed the homeostasis of mature DCs or pre-DCs in the lung, dermis, and spleen. CD11c-cre-Irf4(-/-) mice selectively lacked a lung-resident CD11c(hi)CD11b(+)SIRPα(+)CD24(+) DC subset, but not other lung CD11b(+) DCs or alveolar macrophages. Numbers of CD11b(+)CD4(+) splenic DCs, but not CD11b(+) dermal DCs, were reduced, indicating cDC2s in the lung and dermis develop via different pathways. Irf4 deficiency did not alter numbers of cDC1s. CD11c-cre-Irf8(-/-) mice lacked lung-resident CD103(+) DCs and splenic CD8α(+) DCs, yet harbored increased IRF4-dependent DCs. This correlated with a reduced number of Irf8(-/-) pre-cDCs, which contained elevated IRF4, suggesting that Irf8 deficiency diverts pre-cDC fate. Analyses of Irf4 and Irf8 haploinsufficient mice showed that, although one Irf4 allele was sufficient for lung cDC2 development, two functional Irf8 alleles were required for differentiation of lung cDC1s. Thus, IRF8 and IRF4 act in pre-cDCs to direct the terminal differentiation of cDC1 and cDC2 subsets in the lung and spleen. These data suggest that variation in IRF4 or IRF8 levels resulting from genetic polymorphisms or environmental cues will govern tissue DC numbers and, therefore, regulate the magnitude of DC functional responses.
Copyright © 2016 by The American Association of Immunologists, Inc.
Figures
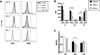


References
-
- Lambrecht BN, Hammad H. Lung dendritic cells in respiratory viral infection and asthma: from protection to immunopathology. Annu Rev Immunol. 2012;30:243–270. - PubMed
-
- Belz GT, Nutt SL. Transcriptional programming of the dendritic cell network. Nat Rev Immunol. 2012;12:101–113. - PubMed
-
- Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek KG, Helft J, Hashimoto D, Chow A, Price J, Greter M, Bogunovic M, Bellemare-Pelletier A, Frenette PS, Randolph GJ, Turley SJ, Merad M. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

