Association of Arrhythmia-Related Genetic Variants With Phenotypes Documented in Electronic Medical Records
- PMID: 26746457
- PMCID: PMC4758131
- DOI: 10.1001/jama.2015.17701
Association of Arrhythmia-Related Genetic Variants With Phenotypes Documented in Electronic Medical Records
Abstract
Importance: Large-scale DNA sequencing identifies incidental rare variants in established Mendelian disease genes, but the frequency of related clinical phenotypes in unselected patient populations is not well established. Phenotype data from electronic medical records (EMRs) may provide a resource to assess the clinical relevance of rare variants.
Objective: To determine the clinical phenotypes from EMRs for individuals with variants designated as pathogenic by expert review in arrhythmia susceptibility genes.
Design, setting, and participants: This prospective cohort study included 2022 individuals recruited for nonantiarrhythmic drug exposure phenotypes from October 5, 2012, to September 30, 2013, for the Electronic Medical Records and Genomics Network Pharmacogenomics project from 7 US academic medical centers. Variants in SCN5A and KCNH2, disease genes for long QT and Brugada syndromes, were assessed for potential pathogenicity by 3 laboratories with ion channel expertise and by comparison with the ClinVar database. Relevant phenotypes were determined from EMRs, with data available from 2002 (or earlier for some sites) through September 10, 2014.
Exposures: One or more variants designated as pathogenic in SCN5A or KCNH2.
Main outcomes and measures: Arrhythmia or electrocardiographic (ECG) phenotypes defined by International Classification of Diseases, Ninth Revision (ICD-9) codes, ECG data, and manual EMR review.
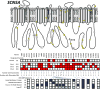
Results: Among 2022 study participants (median age, 61 years [interquartile range, 56-65 years]; 1118 [55%] female; 1491 [74%] white), a total of 122 rare (minor allele frequency <0.5%) nonsynonymous and splice-site variants in 2 arrhythmia susceptibility genes were identified in 223 individuals (11% of the study cohort). Forty-two variants in 63 participants were designated potentially pathogenic by at least 1 laboratory or ClinVar, with low concordance across laboratories (Cohen κ = 0.26). An ICD-9 code for arrhythmia was found in 11 of 63 (17%) variant carriers vs 264 of 1959 (13%) of those without variants (difference, +4%; 95% CI, -5% to +13%; P = .35). In the 1270 (63%) with ECGs, corrected QT intervals were not different in variant carriers vs those without (median, 429 vs 439 milliseconds; difference, -10 milliseconds; 95% CI, -16 to +3 milliseconds; P = .17). After manual review, 22 of 63 participants (35%) with designated variants had any ECG or arrhythmia phenotype, and only 2 had corrected QT interval longer than 500 milliseconds.
Conclusions and relevance: Among laboratories experienced in genetic testing for cardiac arrhythmia disorders, there was low concordance in designating SCN5A and KCNH2 variants as pathogenic. In an unselected population, the putatively pathogenic genetic variants were not associated with an abnormal phenotype. These findings raise questions about the implications of notifying patients of incidental genetic findings.
Figures

Comment in
-
Telling patients of incidental genetic findings is questionable, researchers warn.BMJ. 2016 Jan 5;352:h7034. doi: 10.1136/bmj.h7034. BMJ. 2016. PMID: 26738744 No abstract available.
-
Establishing the Clinical Validity of Arrhythmia-Related Genetic Variations Using the Electronic Medical Record: A Valid Take on Precision Medicine?JAMA. 2016 Jan 5;315(1):33-5. doi: 10.1001/jama.2015.17702. JAMA. 2016. PMID: 26746455 No abstract available.
-
Arrhythmias: Opening Pandora's Box -- incidental genetic findings.Nat Rev Cardiol. 2016 Apr;13(4):187-8. doi: 10.1038/nrcardio.2016.27. Epub 2016 Mar 3. Nat Rev Cardiol. 2016. PMID: 26935036
-
Long QT Syndrome and Potentially Pathogenic Genetic Variants.JAMA. 2016 Jun 14;315(22):2467. doi: 10.1001/jama.2016.2918. JAMA. 2016. PMID: 27299623 No abstract available.
-
Long QT Syndrome and Potentially Pathogenic Genetic Variants--In Reply.JAMA. 2016 Jun 14;315(22):2467-8. doi: 10.1001/jama.2016.2921. JAMA. 2016. PMID: 27299624 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- UL1 TR000427/TR/NCATS NIH HHS/United States
- R01 AG034676/AG/NIA NIH HHS/United States
- U01GM097119/GM/NIGMS NIH HHS/United States
- U01 HG006380/HG/NHGRI NIH HHS/United States
- U01HG006388/HG/NHGRI NIH HHS/United States
- R01 CA138461/CA/NCI NIH HHS/United States
- U19 GM061388/GM/NIGMS NIH HHS/United States
- U01HG006379/HG/NHGRI NIH HHS/United States
- U01 HG006385/HG/NHGRI NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- U01HG004438/HG/NHGRI NIH HHS/United States
- U01 HG006375/HG/NHGRI NIH HHS/United States
- U01HG006375/HG/NHGRI NIH HHS/United States
- U01 HG004438/HG/NHGRI NIH HHS/United States
- UL1TR000445/TR/NCATS NIH HHS/United States
- T32 GM007454/GM/NIGMS NIH HHS/United States
- U01 HG008657/HG/NHGRI NIH HHS/United States
- U01HG006830/HG/NHGRI NIH HHS/United States
- U01 HG006382/HG/NHGRI NIH HHS/United States
- U01HG006382/HG/NHGRI NIH HHS/United States
- R01 GM028157/GM/NIGMS NIH HHS/United States
- U01 HG006389/HG/NHGRI NIH HHS/United States
- U01AG006781/AG/NIA NIH HHS/United States
- U01 HG008672/HG/NHGRI NIH HHS/United States
- U19HL069757/HL/NHLBI NIH HHS/United States
- U01 HG008684/HG/NHGRI NIH HHS/United States
- UL1TR000427/TR/NCATS NIH HHS/United States
- U01 HG006828/HG/NHGRI NIH HHS/United States
- U01HG006380/HG/NHGRI NIH HHS/United States
- U01 AG006781/AG/NIA NIH HHS/United States
- U01 HG005137/HG/NHGRI NIH HHS/United States
- K23 GM104401/GM/NIGMS NIH HHS/United States
- U01 HG006388/HG/NHGRI NIH HHS/United States
- U01 HG006378/HG/NHGRI NIH HHS/United States
- U19 HL069757/HL/NHLBI NIH HHS/United States
- R01AG034676/AG/NIA NIH HHS/United States
- U01 HG008673/HG/NHGRI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- U19GM61388/GM/NIGMS NIH HHS/United States
- U01HG006385/HG/NHGRI NIH HHS/United States
- U01HG006378/HG/NHGRI NIH HHS/United States
- F30 HL127904/HL/NHLBI NIH HHS/United States
- U01HG005137/HG/NHGRI NIH HHS/United States
- T32 GM082773/GM/NIGMS NIH HHS/United States
- U01 HG008701/HG/NHGRI NIH HHS/United States
- U01HG006828/HG/NHGRI NIH HHS/United States
- R01CA138461/CA/NCI NIH HHS/United States
- U01HG006389/HG/NHGRI NIH HHS/United States
- R01GM28157/GM/NIGMS NIH HHS/United States
- U01 HG006830/HG/NHGRI NIH HHS/United States
- U01 GM097119/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

