Genome-wide binding analysis of the transcriptional regulator TrmBL1 in Pyrococcus furiosus
- PMID: 26747700
- PMCID: PMC4706686
- DOI: 10.1186/s12864-015-2360-0
Genome-wide binding analysis of the transcriptional regulator TrmBL1 in Pyrococcus furiosus
Abstract
Background: Several in vitro studies document the function of the transcriptional regulator TrmBL1 of Pyrococcus furiosus. These data indicate that the protein can act as repressor or activator and is mainly involved in transcriptional control of sugar uptake and in the switch between glycolysis and gluconeogenesis. The aim of this study was to complement the in vitro data with an in vivo analysis using ChIP-seq to explore the genome-wide binding profile of TrmBL1 under glycolytic and gluconeogenic growth conditions.
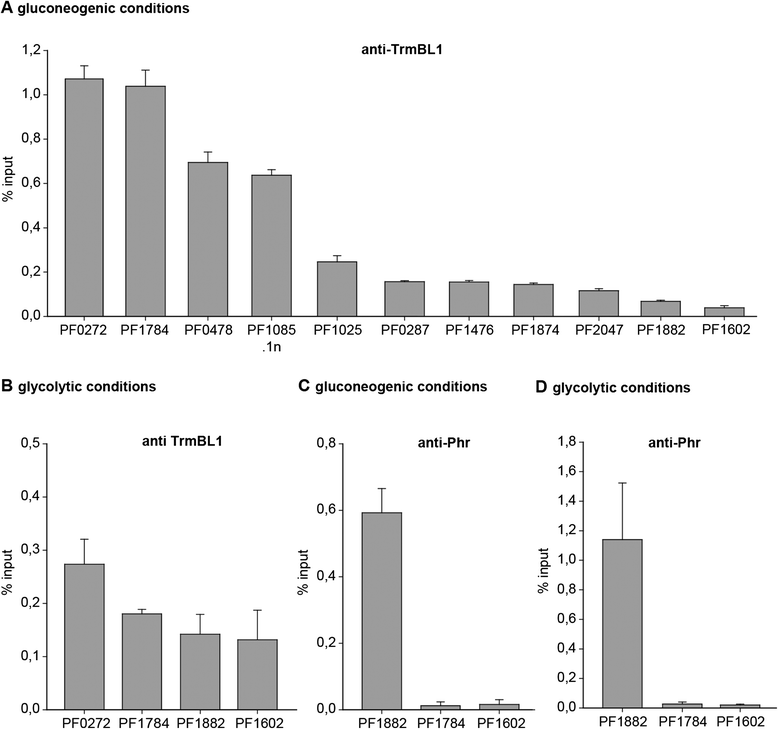
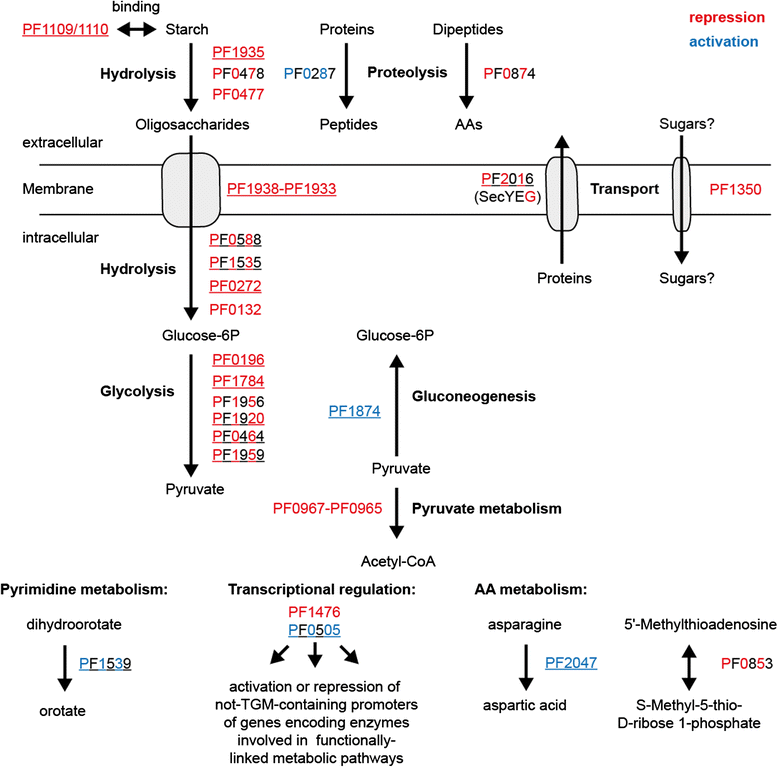
Results: The ChIP-seq analysis revealed under gluconeogenic growth conditions 28 TrmBL1 binding sites where the TGM is located upstream of coding regions and no binding sites under glycolytic conditions. The experimental confirmation of the binding sites using qPCR, EMSA, DNase I footprinting and in vitro transcription experiments validated the in vivo identified TrmBL1 binding sites. Furthermore, this study provides evidence that TrmBL1 is also involved in transcriptional regulation of additional cellular processes e.g. amino acid metabolism, transcriptional control or metabolic pathways. In the initial setup we were interested to include the binding analysis of TrmB, an additional member of the TrmB family, but western blot experiments and the ChIP-seq data indicated that the corresponding gene is deleted in our Pyrococcus strain. A detailed analysis of a new type strain demonstrated that a 16 kb fragment containing the trmb gene is almost completely deleted after the first re-cultivation.
Conclusions: The identified binding sites in the P. furiosus genome classified TrmBL1 as a more global regulator as hitherto known. Furthermore, the high resolution of the mapped binding positions enabled reliable predictions, if TrmBL1 activates (binding site upstream of the promoter) or represses transcription (binding site downstream) of the corresponding genes.
Figures






References
-
- Garrett RA, Klenk H. Archaea: Evolution, physiology, and molecular biology. Malden, MA: Blackwell Pub; 2007.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

