Systematic Phenomics Analysis Deconvolutes Genes Mutated in Intellectual Disability into Biologically Coherent Modules
- PMID: 26748517
- PMCID: PMC4716705
- DOI: 10.1016/j.ajhg.2015.11.024
Systematic Phenomics Analysis Deconvolutes Genes Mutated in Intellectual Disability into Biologically Coherent Modules
Abstract
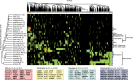
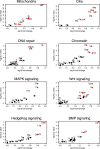
Intellectual disability (ID) disorders are genetically and phenotypically extremely heterogeneous. Can this complexity be depicted in a comprehensive way as a means of facilitating the understanding of ID disorders and their underlying biology? We provide a curated database of 746 currently known genes, mutations in which cause ID (ID-associated genes [ID-AGs]), classified according to ID manifestation and associated clinical features. Using this integrated resource, we show that ID-AGs are substantially enriched with co-expression, protein-protein interactions, and specific biological functions. Systematic identification of highly enriched functional themes and phenotypes revealed typical phenotype combinations characterizing process-defined groups of ID disorders, such as chromatin-related disorders and deficiencies in DNA repair. Strikingly, phenotype classification efficiently breaks down ID-AGs into subsets with significantly elevated biological coherence and predictive power. Custom-made functional Drosophila datasets revealed further characteristic phenotypes among ID-AGs and specific clinical classes. Our study and resource provide systematic insights into the molecular and clinical landscape of ID disorders, represent a significant step toward overcoming current limitations in ID research, and prove the utility of systematic human and cross-species phenomics analyses in highly heterogeneous genetic disorders.
Copyright © 2016 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Schalock R.L., Borthwick-Duffy S.A., Bradley V.J., Buntinx W.H.E., Coulter D.L., Craig E.M., Gomez S.C., Lachapelle Y., Luckasson R., Reeve A. American Association on Intellectual and Developmental Disabilities; 2010. Intellectual Disability: Definition, Classification, and Systems of Supports.
-
- Ropers H.H. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 2010;11:161–187. - PubMed
-
- Grozeva D., Carss K., Spasic-Boskovic O., Tejada M.I., Gecz J., Shaw M., Corbett M., Haan E., Thompson E., Friend K., Italian X-linked Mental Retardation Project. UK10K Consortium. GOLD Consortium Targeted Next-Generation Sequencing Analysis of 1,000 Individuals with Intellectual Disability. Hum. Mutat. 2015;36:1197–1204. - PMC - PubMed
-
- Bahi-Buisson N., Poirier K., Fourniol F., Saillour Y., Valence S., Lebrun N., Hully M., Bianco C.F., Boddaert N., Elie C., LIS-Tubulinopathies Consortium The wide spectrum of tubulinopathies: what are the key features for the diagnosis? Brain. 2014;137:1676–1700. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

