Simultaneous Multiplexed Measurement of RNA and Proteins in Single Cells
- PMID: 26748716
- PMCID: PMC4713867
- DOI: 10.1016/j.celrep.2015.12.021
Simultaneous Multiplexed Measurement of RNA and Proteins in Single Cells
Abstract
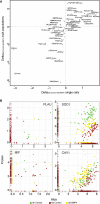
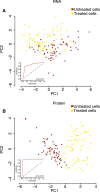
Significant advances have been made in methods to analyze genomes and transcriptomes of single cells, but to fully define cell states, proteins must also be accessed as central actors defining a cell's phenotype. Methods currently used to analyze endogenous protein expression in single cells are limited in specificity, throughput, or multiplex capability. Here, we present an approach to simultaneously and specifically interrogate large sets of protein and RNA targets in lysates from individual cells, enabling investigations of cell functions and responses. We applied our method to investigate the effects of BMP4, an experimental therapeutic agent, on early-passage glioblastoma cell cultures. We uncovered significant heterogeneity in responses to treatment at levels of RNA and protein, with a subset of cells reacting in a distinct manner to BMP4. Moreover, we found overall poor correlation between protein and RNA at the level of single cells, with proteins more accurately defining responses to treatment.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

