Material Cues as Potent Regulators of Epigenetics and Stem Cell Function
- PMID: 26748755
- PMCID: PMC5409508
- DOI: 10.1016/j.stem.2015.12.012
Material Cues as Potent Regulators of Epigenetics and Stem Cell Function
Abstract
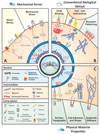
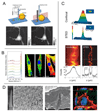
Biophysical signals act as potent regulators of stem cell function, lineage commitment, and epigenetic status. In recent years, synthetic biomaterials have been used to study a wide range of outside-in signaling events, and it is now well appreciated that material cues modulate the epigenome. Here, we review the role of extracellular signals in guiding stem cell behavior via epigenetic regulation, and we stress the role of physicochemical material properties as an often-overlooked modulator of intracellular signaling. We also highlight promising new research tools for ongoing interrogation of the stem cell-material interface.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
-
- Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol. 2014;16:281–293. - PMC - PubMed
-
- Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol. 2004;22:863–866. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

