No longer a nuisance: long non-coding RNAs join CENP-A in epigenetic centromere regulation
- PMID: 26748759
- PMCID: PMC11108473
- DOI: 10.1007/s00018-015-2124-7
No longer a nuisance: long non-coding RNAs join CENP-A in epigenetic centromere regulation
Abstract
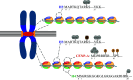
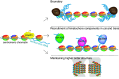
Centromeres represent the basis for kinetochore formation, and are essential for proper chromosome segregation during mitosis. Despite these essential roles, centromeres are not defined by specific DNA sequences, but by epigenetic means. The histone variant CENP-A controls centromere identity epigenetically and is essential for recruiting kinetochore components that attach the chromosomes to the mitotic spindle during mitosis. Recently, a new player in centromere regulation has emerged: long non-coding RNAs transcribed from repetitive regions of centromeric DNA function in regulating centromeres epigenetically. This review summarizes recent findings on the essential roles that transcription, pericentromeric transcripts, and centromere-derived RNAs play in centromere biology.
Keywords: Centromere; Epigenetics; Kinetochore; Long non-coding RNA; Mitosis; Transcription.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

