Seedlings Transduce the Depth and Mechanical Pressure of Covering Soil Using COP1 and Ethylene to Regulate EBF1/EBF2 for Soil Emergence
- PMID: 26748855
- PMCID: PMC5108888
- DOI: 10.1016/j.cub.2015.11.053
Seedlings Transduce the Depth and Mechanical Pressure of Covering Soil Using COP1 and Ethylene to Regulate EBF1/EBF2 for Soil Emergence
Abstract
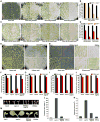
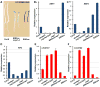
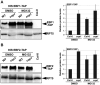
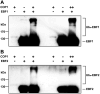
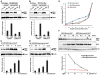
The survival of seed plants in natural environments requires the successful emergence from the soil. In this process, the ethylene signaling pathway is utilized by plants to sense and respond to the mechanical resistance of the soil. Here, we report that constitutive photomorphogenesis 1 (COP1), a central repressor of light signaling, is a key component required for seedlings to sense the depth of soil overlay. Mutation in COP1 causes severe defects in penetrating soil, due to decreased level of EIN3, a master transcription factor in ethylene pathway that mediates seedling emergence. We show that COP1 directly targets the F box proteins EBF1 and EBF2 for ubiquitination and degradation, thus stabilizing EIN3. As seedlings grow toward the surface, the depth of soil overlay decreases, resulting in a gradual increase of light fluences. COP1 channels the light signals, while ethylene transduces the information on soil mechanical conditions, which cooperatively control EIN3 protein levels to promote seedling emergence from the soil. The COP1-EBF1/2-EIN3 module reveals a mechanism by which plants sense the depth to surface and uncovers a novel regulatory paradigm of an ubiquitin E3 ligase cascade.
Keywords: COP1; EBF1 and EBF2; EIN3; ethylene signaling; seedling emergence.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

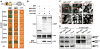
Comment in
-
Plant Biology: Seedling Emergence through Soil.Curr Biol. 2016 Jan 25;26(2):R68-R70. doi: 10.1016/j.cub.2015.12.003. Curr Biol. 2016. PMID: 26811891
-
Seedling signalling: Ubiquitin ligases acting in tandem.Nat Plants. 2016 Feb 3;2:16001. doi: 10.1038/nplants.2016.1. Nat Plants. 2016. PMID: 27249197 No abstract available.
-
Stabilizing the Transcription Factors by E3 Ligase COP1.Trends Plant Sci. 2017 Dec;22(12):999-1001. doi: 10.1016/j.tplants.2017.09.012. Epub 2017 Oct 4. Trends Plant Sci. 2017. PMID: 28988633
Similar articles
-
The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling.Plant Cell. 2007 Feb;19(2):509-23. doi: 10.1105/tpc.106.048140. Epub 2007 Feb 16. Plant Cell. 2007. PMID: 17307926 Free PMC article.
-
The Red Light Receptor Phytochrome B Directly Enhances Substrate-E3 Ligase Interactions to Attenuate Ethylene Responses.Dev Cell. 2016 Dec 5;39(5):597-610. doi: 10.1016/j.devcel.2016.10.020. Epub 2016 Nov 23. Dev Cell. 2016. PMID: 27889482 Free PMC article.
-
EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2.Cell. 2003 Dec 12;115(6):679-89. doi: 10.1016/s0092-8674(03)00968-1. Cell. 2003. PMID: 14675533
-
Paradigms and paradox in the ethylene signaling pathway and interaction network.Mol Plant. 2011 Jul;4(4):626-34. doi: 10.1093/mp/ssr042. Epub 2011 Jun 20. Mol Plant. 2011. PMID: 21690206 Review.
-
Genetic basis of ethylene perception and signal transduction in Arabidopsis.J Integr Plant Biol. 2008 Jul;50(7):808-15. doi: 10.1111/j.1744-7909.2008.00710.x. J Integr Plant Biol. 2008. PMID: 18713391 Review.
Cited by
-
SDC mediates DNA methylation-controlled clock pace by interacting with ZTL in Arabidopsis.Nucleic Acids Res. 2021 Apr 19;49(7):3764-3780. doi: 10.1093/nar/gkab128. Nucleic Acids Res. 2021. PMID: 33675668 Free PMC article.
-
Ethylene Signaling Facilitates Plant Adaption to Physical Barriers.Front Plant Sci. 2021 Jul 29;12:697988. doi: 10.3389/fpls.2021.697988. eCollection 2021. Front Plant Sci. 2021. PMID: 34394151 Free PMC article. Review.
-
Phosphorylation status of Bβ subunit acts as a switch to regulate the function of phosphatase PP2A in ethylene-mediated root growth inhibition.New Phytol. 2022 Dec;236(5):1762-1778. doi: 10.1111/nph.18467. Epub 2022 Sep 26. New Phytol. 2022. PMID: 36073540 Free PMC article.
-
Overexpression of Lectin Receptor-Like Kinase 1 in Tomato Confers Resistance to Fusarium oxysporum f. sp. Radicis-Lycopersici.Front Plant Sci. 2022 Feb 3;13:836269. doi: 10.3389/fpls.2022.836269. eCollection 2022. Front Plant Sci. 2022. PMID: 35185997 Free PMC article.
-
Coordinated Shoot and Root Responses to Light Signaling in Arabidopsis.Plant Commun. 2020 Jan 22;1(2):100026. doi: 10.1016/j.xplc.2020.100026. eCollection 2020 Mar 9. Plant Commun. 2020. PMID: 33367230 Free PMC article. Review.
References
-
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. - PubMed
-
- Von Arnim A, Deng XW. Light Control of Seedling Development. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:215–243. - PubMed
-
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annual Review of Genetics. 2004;38:87–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

