Interleukin-2-Dependent Allergen-Specific Tissue-Resident Memory Cells Drive Asthma
- PMID: 26750312
- PMCID: PMC4720536
- DOI: 10.1016/j.immuni.2015.11.004
Interleukin-2-Dependent Allergen-Specific Tissue-Resident Memory Cells Drive Asthma
Abstract
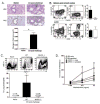
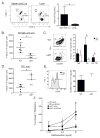
Exposure to inhaled allergens generates T helper 2 (Th2) CD4(+) T cells that contribute to episodes of inflammation associated with asthma. Little is known about allergen-specific Th2 memory cells and their contribution to airway inflammation. We generated reagents to understand how endogenous CD4(+) T cells specific for a house dust mite (HDM) allergen form and function. After allergen exposure, HDM-specific memory cells persisted as central memory cells in the lymphoid organs and tissue-resident memory cells in the lung. Experimental blockade of lymphocyte migration demonstrated that lung-resident cells were sufficient to induce airway hyper-responsiveness, which depended upon CD4(+) T cells. Investigation into the differentiation of pathogenic Trm cells revealed that interleukin-2 (IL-2) signaling was required for residency and directed a program of tissue homing migrational cues. These studies thus identify IL-2-dependent resident Th2 memory cells as drivers of lung allergic responses.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Location, Location, Location: Localized Memory Cells Take Residence in the Allergic Lung.Immunity. 2016 Jan 19;44(1):13-15. doi: 10.1016/j.immuni.2015.12.012. Immunity. 2016. PMID: 26789918
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

