CFTR is a tumor suppressor gene in murine and human intestinal cancer
- PMID: 26751771
- PMCID: PMC4940277
- DOI: 10.1038/onc.2015.483
CFTR is a tumor suppressor gene in murine and human intestinal cancer
Erratum in
-
CFTR is a tumor suppressor gene in murine and human intestinal cancer.Oncogene. 2017 Jun 15;36(24):3504. doi: 10.1038/onc.2017.3. Epub 2017 Feb 13. Oncogene. 2017. PMID: 28192405
Abstract
CFTR, the cystic fibrosis (CF) gene, encodes for the CFTR protein that plays an essential role in anion regulation and tissue homeostasis of various epithelia. In the gastrointestinal (GI) tract CFTR promotes chloride and bicarbonate secretion, playing an essential role in ion and acid-base homeostasis. Cftr has been identified as a candidate driver gene for colorectal cancer (CRC) in several Sleeping Beauty DNA transposon-based forward genetic screens in mice. Further, recent epidemiological and clinical studies indicate that CF patients are at high risk for developing tumors in the colon. To investigate the effects of CFTR dysregulation on GI cancer, we generated Apc(Min) mice that carried an intestinal-specific knockout of Cftr. Our results indicate that Cftr is a tumor suppressor gene in the intestinal tract as Cftr mutant mice developed significantly more tumors in the colon and the entire small intestine. In Apc(+/+) mice aged to ~1 year, Cftr deficiency alone caused the development of intestinal tumors in >60% of mice. Colon organoid formation was significantly increased in organoids created from Cftr mutant mice compared with wild-type controls, suggesting a potential role of Cftr in regulating the intestinal stem cell compartment. Microarray data from the Cftr-deficient colon and the small intestine identified dysregulated genes that belong to groups of immune response, ion channel, intestinal stem cell and other growth signaling regulators. These associated clusters of genes were confirmed by pathway analysis using Ingenuity Pathway Analysis and gene set enrichment analysis (GSEA). We also conducted RNA Seq analysis of tumors from Apc(+/+) Cftr knockout mice and identified sets of genes dysregulated in tumors including altered Wnt β-catenin target genes. Finally we analyzed expression of CFTR in early stage human CRC patients stratified by risk of recurrence and found that loss of expression of CFTR was significantly associated with poor disease-free survival.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


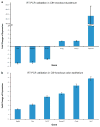
References
-
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. - PubMed
-
- Pedersen SF, Stock C. Ion channels and transporters in cancer: pathophysiology, regulation, and clinical potential. Cancer Res. 2013;73:1658–1661. - PubMed
-
- Neglia JP, FitzSimmons SC, Maisonneuve P, Schöni MH, Schöni-Affolter F, Corey M, et al. The risk of cancer among patients with cystic fibrosis. Cystic Fibrosis and Cancer Study Group. N Engl J Med. 1995;332:494–499. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

