Genome-Wide Analysis Reveals Novel Regulators of Growth in Drosophila melanogaster
- PMID: 26751788
- PMCID: PMC4709145
- DOI: 10.1371/journal.pgen.1005616
Genome-Wide Analysis Reveals Novel Regulators of Growth in Drosophila melanogaster
Abstract
Organismal size depends on the interplay between genetic and environmental factors. Genome-wide association (GWA) analyses in humans have implied many genes in the control of height but suffer from the inability to control the environment. Genetic analyses in Drosophila have identified conserved signaling pathways controlling size; however, how these pathways control phenotypic diversity is unclear. We performed GWA of size traits using the Drosophila Genetic Reference Panel of inbred, sequenced lines. We find that the top associated variants differ between traits and sexes; do not map to canonical growth pathway genes, but can be linked to these by epistasis analysis; and are enriched for genes and putative enhancers. Performing GWA on well-studied developmental traits under controlled conditions expands our understanding of developmental processes underlying phenotypic diversity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
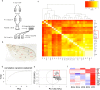

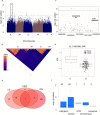
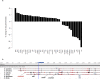
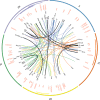
Comment in
-
The Mighty Fruit Fly Moves into Outbred Genetics.PLoS Genet. 2016 Nov 3;12(11):e1006388. doi: 10.1371/journal.pgen.1006388. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27812108 Free PMC article. No abstract available.
References
-
- Johnston LA, Gallant P. Control of growth and organ size in Drosophila. Bioessays 2002; 24: 54–64. - PubMed
-
- Mirth CK, Riddiford LM. Size assessment and growth control: how adult size is determined in insects. Bioessays 2007; 29: 344–355. - PubMed
-
- Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol.2003; 13: 79–85. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

