KIF1A inhibition immortalizes brain stem cells but blocks BDNF-mediated neuronal migration
- PMID: 26752160
- PMCID: PMC4731285
- DOI: 10.1038/nn.4213
KIF1A inhibition immortalizes brain stem cells but blocks BDNF-mediated neuronal migration
Abstract
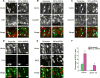
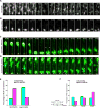
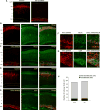
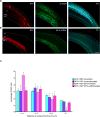
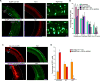
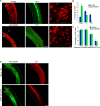
Brain neural stem cells (radial glial progenitors, RGPs) undergo a mysterious form of cell cycle-entrained interkinetic nuclear migration (INM) that is driven apically by cytoplasmic dynein and basally by the kinesin KIF1A, which has recently been implicated in human brain developmental disease. To understand the consequences of altered basal INM and the roles of KIF1A in disease, we performed constitutive and conditional RNAi and expressed mutant KIF1A in E16 to P7 rat RGPs and neurons. RGPs inhibited in basal INM still showed normal cell cycle progression, although neurogenic divisions were severely reduced. Postmitotic neuronal migration was independently disrupted at the multipolar stage and accompanied by premature ectopic expression of neuronal differentiation markers. Similar effects were unexpectedly observed throughout the layer of surrounding control cells, mimicked by Bdnf (brain-derived neurotrophic factor) or Dcx RNAi, and rescued by BDNF application. These results identify sequential and independent roles for KIF1A and provide an important new approach for reversing the effects of human disease.
Conflict of interest statement
Conflict of interest statement. None declared
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

