Cucumber Possesses a Single Terminal Alternative Oxidase Gene That is Upregulated by Cold Stress and in the Mosaic (MSC) Mitochondrial Mutants
- PMID: 26752808
- PMCID: PMC4695503
- DOI: 10.1007/s11105-015-0883-9
Cucumber Possesses a Single Terminal Alternative Oxidase Gene That is Upregulated by Cold Stress and in the Mosaic (MSC) Mitochondrial Mutants
Abstract
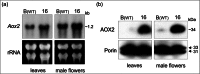
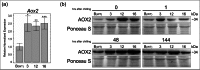
Alternative oxidase (AOX) is a mitochondrial terminal oxidase which is responsible for an alternative route of electron transport in the respiratory chain. This nuclear-encoded enzyme is involved in a major path of survival under adverse conditions by transfer of electrons from ubiquinol instead of the main cytochrome pathway. AOX protects against unexpected inhibition of the cytochrome c oxidase pathway and plays an important role in stress tolerance. Two AOX subfamilies (AOX1 and AOX2) exist in higher plants and are usually encoded by small gene families. In this study, genome-wide searches and cloning were completed to identify and characterize AOX genes in cucumber (Cucumis sativus L.). Our results revealed that cucumber possesses no AOX1 gene(s) and only a single AOX2 gene located on chromosome 4. Expression studies showed that AOX2 in wild-type cucumber is constitutively expressed at low levels and is upregulated by cold stress. AOX2 transcripts and protein were detected in leaves and flowers of wild-type plants, with higher levels in the three independently derived mosaic (MSC) mitochondrial mutants. Because cucumber possesses a single AOX gene and its expression increases under cold stress and in the MSC mutants, this plant is a unique and intriguing model to study AOX expression and regulation particularly in the context of mitochondria-to-nucleus retrograde signaling.
Keywords: Alternative oxidase; Cucumis sativus; Gene structure; Mitochondrial mutants.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
