Erratum to: No training required: experimental tests support homology-based DNA assembly as a best practice in synthetic biology
- PMID: 26752998
- PMCID: PMC4705622
- DOI: 10.1186/s13036-015-0013-0
Erratum to: No training required: experimental tests support homology-based DNA assembly as a best practice in synthetic biology
Abstract
[This corrects the article DOI: 10.1186/s13036-015-0006-z.].
Figures
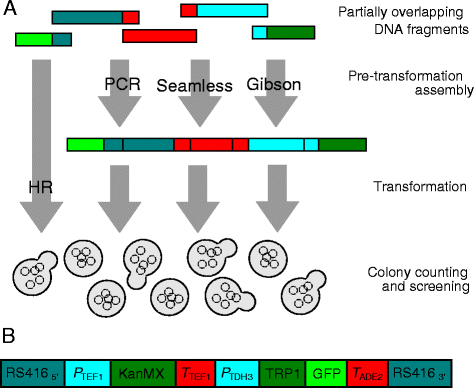
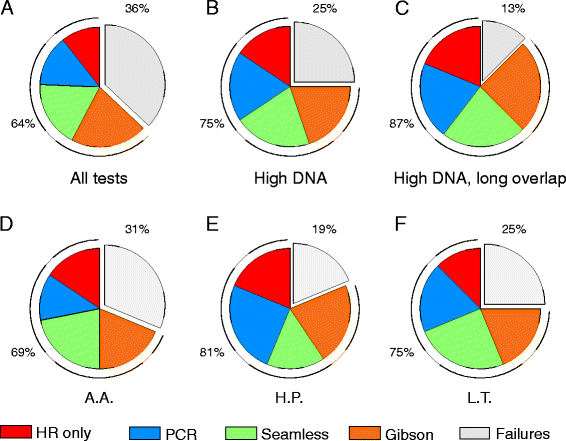
Erratum for
-
No training required: experimental tests support homology-based DNA assembly as a best practice in synthetic biology.J Biol Eng. 2015 Jun 12;9:8. doi: 10.1186/s13036-015-0006-z. eCollection 2015. J Biol Eng. 2015. PMID: 26075023 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

