Favorable prognostic influence of T-box transcription factor Eomesodermin in metastatic renal cell cancer patients
- PMID: 26753694
- PMCID: PMC11029520
- DOI: 10.1007/s00262-015-1786-1
Favorable prognostic influence of T-box transcription factor Eomesodermin in metastatic renal cell cancer patients
Abstract
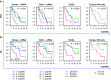
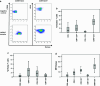
T-box transcription factors, T-box expressed in T cells (T-bet) encoded by Tbx21 and Eomesodermin (Eomes), drive the differentiation of effector/memory T cell lineages and NK cells. The aim of the study was to determine the prognostic influence of the expression of these transcription factors in peripheral blood (pB) in a cohort of 41 metastatic (m) RCC patients before receiving sorafenib treatment and to analyze their association with the immunophenotype in pB. In contrast to Tbx21, in the multivariate analysis including clinical features, Eomes mRNA expression was identified as an independent good prognostic factor for progression-free survival (PFS, p = 0.042) and overall survival (OS, p = 0.001) in addition to a favorable ECOG performance status (p = 0.01 and p = 0.008, respectively). Eomes expression correlated positively not only with expression of Tbx21 and TGFβ1 mRNA, but also with mRNA expression of the activation marker ICOS, and with in vivo activated HLA-DR(+) T cells. Eomes expression was negatively associated with TNFα-producing T cells. On protein level, Eomes was mainly expressed by CD56(+)CD3(-) NK cells in pB. In conclusion, we identified a higher Eomes mRNA expression as an independent good prognostic factor for OS and PFS in mRCC patients treated with sorafenib.
Keywords: Effector T cells; Eomesodermin; Memory T cells; Natural killer cells; Renal cell carcinoma; T-box expressed in T cells.
Conflict of interest statement
All authors declare that they have no conflict of interest including any financial, personal, or other relationships with other people or organizations within that could inappropriately influence their work.
Figures

Similar articles
-
Systemic immune tuning in renal cell carcinoma: favorable prognostic impact of TGF-β1 mRNA expression in peripheral blood mononuclear cells.J Immunother. 2011 Jan;34(1):113-9. doi: 10.1097/CJI.0b013e3181fb6580. J Immunother. 2011. PMID: 21150720
-
Characterization of eomesodermin and T-bet expression by allostimulated CD8+ T cells of healthy volunteers and kidney transplant patients in relation to graft outcome.Clin Exp Immunol. 2018 Nov;194(2):259-272. doi: 10.1111/cei.13162. Epub 2018 Sep 23. Clin Exp Immunol. 2018. PMID: 30246373 Free PMC article.
-
Eomesodermin spatiotemporally orchestrates the early and late stages of NK cell development by targeting KLF2 and T-bet, respectively.Cell Mol Immunol. 2024 Jul;21(7):662-673. doi: 10.1038/s41423-024-01164-8. Epub 2024 May 13. Cell Mol Immunol. 2024. PMID: 38740922 Free PMC article.
-
T-bet and Eomes govern differentiation and function of mouse and human NK cells and ILC1.Eur J Immunol. 2018 May;48(5):738-750. doi: 10.1002/eji.201747299. Epub 2018 Feb 28. Eur J Immunol. 2018. PMID: 29424438 Review.
-
[Therapeutic indication of interferons for renal cell carcinoma].Nihon Rinsho. 2006 Feb;64 Suppl 2:647-51. Nihon Rinsho. 2006. PMID: 16523970 Review. Japanese. No abstract available.
Cited by
-
TIGIT+ TIM-3+ NK cells are correlated with NK cell exhaustion and disease progression in patients with hepatitis B virus‑related hepatocellular carcinoma.Oncoimmunology. 2021 Jun 28;10(1):1942673. doi: 10.1080/2162402X.2021.1942673. eCollection 2021. Oncoimmunology. 2021. PMID: 34249476 Free PMC article.
-
Eomes promotes esophageal carcinoma progression by recruiting Treg cells through the CCL20-CCR6 pathway.Cancer Sci. 2021 Jan;112(1):144-154. doi: 10.1111/cas.14712. Epub 2020 Nov 17. Cancer Sci. 2021. PMID: 33113266 Free PMC article.
-
Identification and Validation of an Immunological Expression-Based Prognostic Signature in Breast Cancer.Front Genet. 2020 Sep 16;11:912. doi: 10.3389/fgene.2020.00912. eCollection 2020. Front Genet. 2020. PMID: 33193571 Free PMC article.
-
Higher Numbers of T-Bet+ Tumor-Infiltrating Lymphocytes Associate with Better Survival in Human Epithelial Ovarian Cancer.Cell Physiol Biochem. 2017;41(2):475-483. doi: 10.1159/000456600. Epub 2017 Jan 30. Cell Physiol Biochem. 2017. PMID: 28214872 Free PMC article.
-
Identification of a seven-gene prognostic model for renal cell carcinoma associated with CD8+T lymphocyte cell.Medicine (Baltimore). 2024 Oct 4;103(40):e39938. doi: 10.1097/MD.0000000000039938. Medicine (Baltimore). 2024. PMID: 39465721 Free PMC article.
References
-
- Busse A, Keilholz U. Immune self-tuning in renal cell carcinoma. Eur Oncol. 2010;6(1):70–75.
-
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6(12):1236–1244. doi: 10.1038/ni1268. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

