Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli
- PMID: 26754917
- PMCID: PMC4728125
- DOI: 10.7554/eLife.07141
Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli
Abstract
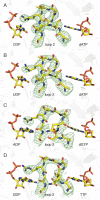
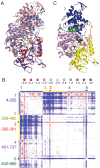
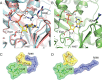
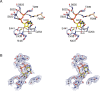
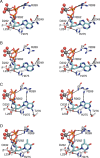
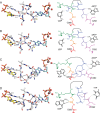
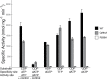
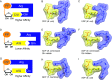
Ribonucleotide reductase (RNR) converts ribonucleotides to deoxyribonucleotides, a reaction that is essential for DNA biosynthesis and repair. This enzyme is responsible for reducing all four ribonucleotide substrates, with specificity regulated by the binding of an effector to a distal allosteric site. In all characterized RNRs, the binding of effector dATP alters the active site to select for pyrimidines over purines, whereas effectors dGTP and TTP select for substrates ADP and GDP, respectively. Here, we have determined structures of Escherichia coli class Ia RNR with all four substrate/specificity effector-pairs bound (CDP/dATP, UDP/dATP, ADP/dGTP, GDP/TTP) that reveal the conformational rearrangements responsible for this remarkable allostery. These structures delineate how RNR 'reads' the base of each effector and communicates substrate preference to the active site by forming differential hydrogen bonds, thereby maintaining the proper balance of deoxynucleotides in the cell.
Keywords: E. coli; X-ray crystallography; allosteric regulation; biochemistry; biophysics; nucleic acid metabolism; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures



References
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D Biological Crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. - DOI - PMC - PubMed
-
- Ahmad MF, Kaushal PS, Wan Q, Wijerathna SR, An X, Huang M, Dealwis CG. Role of arginine 293 and glutamine 288 in communication between catalytic and allosteric sites in yeast ribonucleotide reductase. Journal of Molecular Biology. 2012;419:315–329. doi: 10.1016/j.jmb.2012.03.014. - DOI - PMC - PubMed
-
- Ando N, Brignole EJ, Zimanyi CM, Funk MA, Yokoyama K, Asturias FJ, Stubbe J, Drennan CL. Structural interconversions modulate activity of Escherichia coli ribonucleotide reductase. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21046–21051. doi: 10.1073/pnas.1112715108. - DOI - PMC - PubMed
-
- Bollinger JM, Hangtong W, Ravi N, Hanh Huynh B, Edmondson D, Stubbe J. Use of rapid kinetics methods to study the assembly of the diferric-tyrosyl radical cofactor of Escherichia coli ribonucleotide reductase. Methods in Enzymology. 1995;258:278–303. doi: 10.1016/0076-6879(95)58052-2. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

