Regulation of Antisense Transcription by NuA4 Histone Acetyltransferase and Other Chromatin Regulatory Factors
- PMID: 26755557
- PMCID: PMC4810466
- DOI: 10.1128/MCB.00808-15
Regulation of Antisense Transcription by NuA4 Histone Acetyltransferase and Other Chromatin Regulatory Factors
Abstract
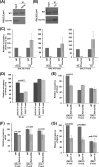
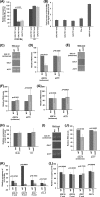
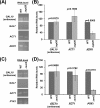
NuA4 histone lysine (K) acetyltransferase (KAT) promotes transcriptional initiation of TATA-binding protein (TBP)-associated factor (TAF)-dependent ribosomal protein genes. TAFs have also been recently found to enhance antisense transcription from the 3' end of the GAL10 coding sequence. However, it remains unknown whether, like sense transcription of the ribosomal protein genes, TAF-dependent antisense transcription of GAL10 also requires NuA4 KAT. Here, we show that NuA4 KAT associates with the GAL10 antisense transcription initiation site at the 3' end of the coding sequence. Such association of NuA4 KAT depends on the Reb1p-binding site that recruits Reb1p activator to the GAL10 antisense transcription initiation site. Targeted recruitment of NuA4 KAT to the GAL10 antisense transcription initiation site promotes GAL10 antisense transcription. Like NuA4 KAT, histone H3 K4/36 methyltransferases and histone H2B ubiquitin conjugase facilitate GAL10 antisense transcription, while the Swi/Snf and SAGA chromatin remodeling/modification factors are dispensable for antisense, but not sense, transcription of GAL10. Taken together, our results demonstrate for the first time the roles of NuA4 KAT and other chromatin regulatory factors in controlling antisense transcription, thus illuminating chromatin regulation of antisense transcription.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Eaf1p Is Required for Recruitment of NuA4 in Targeting TFIID to the Promoters of the Ribosomal Protein Genes for Transcriptional Initiation In Vivo.Mol Cell Biol. 2015 Sep 1;35(17):2947-64. doi: 10.1128/MCB.01524-14. Epub 2015 Jun 22. Mol Cell Biol. 2015. PMID: 26100014 Free PMC article.
-
NuA4 links methylation of histone H3 lysines 4 and 36 to acetylation of histones H4 and H3.J Biol Chem. 2014 Nov 21;289(47):32656-70. doi: 10.1074/jbc.M114.585588. Epub 2014 Oct 9. J Biol Chem. 2014. PMID: 25301943 Free PMC article.
-
NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5.Mol Cell Biol. 2009 Dec;29(24):6473-87. doi: 10.1128/MCB.01033-09. Epub 2009 Oct 12. Mol Cell Biol. 2009. PMID: 19822662 Free PMC article.
-
Share and share alike: the role of Tra1 from the SAGA and NuA4 coactivator complexes.Transcription. 2019 Feb;10(1):37-43. doi: 10.1080/21541264.2018.1530936. Epub 2018 Oct 30. Transcription. 2019. PMID: 30375921 Free PMC article. Review.
-
Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
Cited by
-
Fine-Tuning of FACT by the Ubiquitin Proteasome System in Regulation of Transcriptional Elongation.Mol Cell Biol. 2016 May 16;36(11):1691-703. doi: 10.1128/MCB.01112-15. Print 2016 Jun 1. Mol Cell Biol. 2016. PMID: 27044865 Free PMC article.
-
Proteasomal Regulation of Mammalian SPT16 in Controlling Transcription.Mol Cell Biol. 2021 Mar 24;41(4):e00452-20. doi: 10.1128/MCB.00452-20. Print 2021 Mar 24. Mol Cell Biol. 2021. PMID: 33526453 Free PMC article.
-
Distinct Functions of the Cap-Binding Complex in Stimulation of Nuclear mRNA Export.Mol Cell Biol. 2019 Apr 2;39(8):e00540-18. doi: 10.1128/MCB.00540-18. Print 2019 Apr 15. Mol Cell Biol. 2019. PMID: 30745412 Free PMC article.
-
UPS writes a new saga of SAGA.Biochim Biophys Acta Gene Regul Mech. 2023 Dec;1866(4):194981. doi: 10.1016/j.bbagrm.2023.194981. Epub 2023 Aug 30. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 37657588 Free PMC article. Review.
-
Two Distinct Regulatory Mechanisms of Transcriptional Initiation in Response to Nutrient Signaling.Genetics. 2018 Jan;208(1):191-205. doi: 10.1534/genetics.117.300518. Epub 2017 Nov 15. Genetics. 2018. PMID: 29141908 Free PMC article.
References
-
- Ohhata T, Hoki Y, Sasaki H, Sado T. 2008. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development 135:227–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
