A novel sweet potato potyvirus open reading frame (ORF) is expressed via polymerase slippage and suppresses RNA silencing
- PMID: 26757490
- PMCID: PMC4979677
- DOI: 10.1111/mpp.12366
A novel sweet potato potyvirus open reading frame (ORF) is expressed via polymerase slippage and suppresses RNA silencing
Abstract
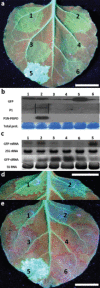
The single-stranded, positive-sense RNA genome of viruses in the genus Potyvirus encodes a large polyprotein that is cleaved to yield 10 mature proteins. The first three cleavage products are P1, HCpro and P3. An additional short open reading frame (ORF), called pipo, overlaps the P3 region of the polyprotein ORF. Four related potyviruses infecting sweet potato (Ipomoea batatas) are predicted to contain a third ORF, called pispo, which overlaps the 3' third of the P1 region. Recently, pipo has been shown to be expressed via polymerase slippage at a conserved GA6 sequence. Here, we show that pispo is also expressed via polymerase slippage at a GA6 sequence, with higher slippage efficiency (∼5%) than at the pipo site (∼1%). Transient expression of recombinant P1 or the 'transframe' product, P1N-PISPO, in Nicotiana benthamiana suppressed local RNA silencing (RNAi), but only P1N-PISPO inhibited short-distance movement of the silencing signal. These results reveal that polymerase slippage in potyviruses is not limited to pipo expression, but can be co-opted for the evolution and expression of further novel gene products.
Keywords: RNA silencing suppression; RNA virus; frameshift; gene expression; overlapping gene; transcriptional slippage.
© 2016 The Authors. Molecular Plant Pathology Published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures






References
-
- Adams, M. , Antoniw, J. and Fauquet, C. (2005) Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 150, 459–479. - PubMed
-
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Angermüller, C. , Biegert, A. and Söding, J. (2012) Discriminative modelling of context‐specific amino acid substitution probabilities. Bioinformatics, 28, 3240–3247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

