Novel Features of Eukaryotic Photosystem II Revealed by Its Crystal Structure Analysis from a Red Alga
- PMID: 26757821
- PMCID: PMC4786707
- DOI: 10.1074/jbc.M115.711689
Novel Features of Eukaryotic Photosystem II Revealed by Its Crystal Structure Analysis from a Red Alga
Abstract
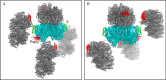
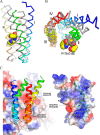
Photosystem II (PSII) catalyzes light-induced water splitting, leading to the evolution of molecular oxygen indispensible for life on the earth. The crystal structure of PSII from cyanobacteria has been solved at an atomic level, but the structure of eukaryotic PSII has not been analyzed. Because eukaryotic PSII possesses additional subunits not found in cyanobacterial PSII, it is important to solve the structure of eukaryotic PSII to elucidate their detailed functions, as well as evolutionary relationships. Here we report the structure of PSII from a red alga Cyanidium caldarium at 2.76 Å resolution, which revealed the structure and interaction sites of PsbQ', a unique, fourth extrinsic protein required for stabilizing the oxygen-evolving complex in the lumenal surface of PSII. The PsbQ' subunit was found to be located underneath CP43 in the vicinity of PsbV, and its structure is characterized by a bundle of four up-down helices arranged in a similar way to those of cyanobacterial and higher plant PsbQ, although helices I and II of PsbQ' were kinked relative to its higher plant counterpart because of its interactions with CP43. Furthermore, two novel transmembrane helices were found in the red algal PSII that are not present in cyanobacterial PSII; one of these helices may correspond to PsbW found only in eukaryotic PSII. The present results represent the first crystal structure of PSII from eukaryotic oxygenic organisms, which were discussed in comparison with the structure of cyanobacterial PSII.
Keywords: crystal structure; crystallography; cyanobacteria; electron transfer complex; membrane protein; photosynthesis; photosystem II; x-ray crystallography.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Zouni A., Witt H. T., Kern J., Fromme P., Krauss N., Saenger W., and Orth P. (2001) Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Angstrom resolution. Nature 409, 739–743 - PubMed
-
- Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., and Iwata S. (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 - PubMed
-
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., and Saenger W. (2009) Cyanobacterial photosystem II at 2.9 Å resolution and role of quinines, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16, 334–342 - PubMed
-
- Umena Y., Kawakami K., Shen J.-R., and Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

