On the Utility of ToxCast™ and ToxPi as Methods for Identifying New Obesogens
- PMID: 26757984
- PMCID: PMC4977052
- DOI: 10.1289/ehp.1510352
On the Utility of ToxCast™ and ToxPi as Methods for Identifying New Obesogens
Abstract
Background: In ToxCast™ Phase I, the U.S. EPA commissioned screening of 320 pesticides, herbicides, fungicides, and other chemicals in a series of high-throughput assays. The agency also developed a toxicological prioritization tool, ToxPi, to facilitate using ToxCast™ assays to predict biological function.
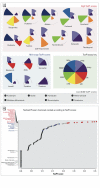
Objectives: We asked whether top-scoring PPARγ activators identified in ToxCast™ Phase I were genuine PPARγ activators and inducers of adipogenesis. Next, we identified ToxCast™ assays that should predict adipogenesis, developed an adipogenesis ToxPi, and asked how well the ToxPi predicted adipogenic activity.
Methods: We used transient transfection to test the ability of ToxCast™ chemicals to modulate PPARγ and RXRα, and differentiation assays employing 3T3-L1 preadipocytes and mouse bone marrow-derived mesenchymal stem cells (mBMSCs) to evaluate the adipogenic capacity of ToxCast™ chemicals.
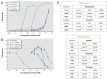
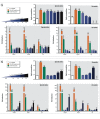
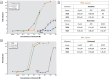
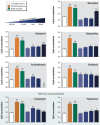
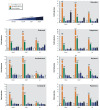
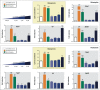
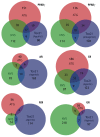
Results: Only 5/21 of the top scoring ToxCast™ PPARγ activators were activators in our assays, 3 were PPARγ antagonists, the remainder were inactive. The bona fide PPARγ activators we identified induced adipogenesis in 3T3-L1 cells and mBMSCs. Only 7 of the 17 chemicals predicted to be active by the ToxPi promoted adipogenesis, 1 inhibited adipogenesis, and 2 of the 7 predicted negatives were also adipogenic. Of these 9 adipogenic chemicals, 3 activated PPARγ, and 1 activated RXRα.
Conclusions: ToxCast™ PPARγ and RXRα assays do not correlate well with laboratory measurements of PPARγ and RXRα activity. The adipogenesis ToxPi performed poorly, perhaps due to the performance of ToxCast™ assays. We observed a modest predictive value of ToxCast™ for PPARγ and RXRα activation and adipogenesis and it is likely that many obesogenic chemicals remain to be identified.
Citation: Janesick AS, Dimastrogiovanni G, Vanek L, Boulos C, Chamorro-García R, Tang W, Blumberg B. 2016. On the utility of ToxCast™ and ToxPi as methods for identifying new obesogens. Environ Health Perspect 124:1214-1226; http://dx.doi.org/10.1289/ehp.1510352.
Conflict of interest statement
The other authors declare they have no actual or potential competing financial interests.
Figures
Comment in
-
Comment on "On the Utility of ToxCast™ and ToxPi as Methods for Identifying New Obesogens".Environ Health Perspect. 2017 Jan 1;125(1):A8-A11. doi: 10.1289/EHP881. Environ Health Perspect. 2017. PMID: 28055944 Free PMC article. No abstract available.
-
Reply to "Comment on 'On the Utility of ToxCast™ and ToxPi as Methods for Identifying New Obesogens'".Environ Health Perspect. 2017 Jan 1;125(1):A12-A14. doi: 10.1289/EHP1122. Environ Health Perspect. 2017. PMID: 28055946 Free PMC article. No abstract available.
Similar articles
-
Prioritizing Environmental Chemicals for Obesity and Diabetes Outcomes Research: A Screening Approach Using ToxCast™ High-Throughput Data.Environ Health Perspect. 2016 Aug;124(8):1141-54. doi: 10.1289/ehp.1510456. Epub 2016 Mar 15. Environ Health Perspect. 2016. PMID: 26978842 Free PMC article.
-
Using ToxCast™ Data to Reconstruct Dynamic Cell State Trajectories and Estimate Toxicological Points of Departure.Environ Health Perspect. 2016 Jul;124(7):910-9. doi: 10.1289/ehp.1409029. Epub 2015 Oct 16. Environ Health Perspect. 2016. PMID: 26473631 Free PMC article.
-
Consideration of dosimetry in evaluation of ToxCast™ data.J Appl Toxicol. 2011 Nov;31(8):741-51. doi: 10.1002/jat.1626. Epub 2011 Mar 5. J Appl Toxicol. 2011. PMID: 21381051
-
On the Utility of ToxCast-Based Predictive Models to Evaluate Potential Metabolic Disruption by Environmental Chemicals.Environ Health Perspect. 2022 May;130(5):57005. doi: 10.1289/EHP6779. Epub 2022 May 9. Environ Health Perspect. 2022. PMID: 35533074 Free PMC article. Review.
-
Environmental Obesogens and Their Impact on Susceptibility to Obesity: New Mechanisms and Chemicals.Endocrinology. 2020 Mar 1;161(3):bqaa024. doi: 10.1210/endocr/bqaa024. Endocrinology. 2020. PMID: 32067051 Free PMC article. Review.
Cited by
-
Characterization of Adipogenic Activity of House Dust Extracts and Semi-Volatile Indoor Contaminants in 3T3-L1 Cells.Environ Sci Technol. 2017 Aug 1;51(15):8735-8745. doi: 10.1021/acs.est.7b01788. Epub 2017 Jul 12. Environ Sci Technol. 2017. PMID: 28699343 Free PMC article.
-
Bibliometrics for Social Validation.PLoS One. 2016 Dec 22;11(12):e0168597. doi: 10.1371/journal.pone.0168597. eCollection 2016. PLoS One. 2016. PMID: 28005974 Free PMC article.
-
Incorporating new approach methodologies in toxicity testing and exposure assessment for tiered risk assessment using the RISK21 approach: Case studies on food contact chemicals.Food Chem Toxicol. 2019 Dec;134:110819. doi: 10.1016/j.fct.2019.110819. Epub 2019 Sep 20. Food Chem Toxicol. 2019. PMID: 31545997 Free PMC article.
-
Integrating ToxPi outputs with ArcGIS Dashboards to identify neighborhood threat levels of contaminant transferal during flood events.J Spat Sci. 2023;68(1):57-69. doi: 10.1080/14498596.2021.1891149. Epub 2021 Feb 28. J Spat Sci. 2023. PMID: 36910889 Free PMC article.
-
Obesity III: Obesogen assays: Limitations, strengths, and new directions.Biochem Pharmacol. 2022 May;199:115014. doi: 10.1016/j.bcp.2022.115014. Epub 2022 Apr 5. Biochem Pharmacol. 2022. PMID: 35393121 Free PMC article. Review.
References
-
- Bevington PR, Robinson DK. Boston, MA: McGraw-Hill Education; 2003. Data Reduction and Error Analysis for the Physical Sciences.
-
- Browne P, Judson RS, Casey WM, Kleinstreuer NC, Thomas RS. Screening chemicals for estrogen receptor bioactivity using a computational model. Environ Sci Technol. 2015;49(14):8804–8814. - PubMed
-
- Chamorro-García R, Kirchner S, Li X, Janesick A, Casey SC, Chow C, et al. 2012. Bisphenol A diglycidyl ether induces adipogenic differentiation of multipotent stromal stem cells through a peroxisome proliferator–activated receptor gamma-independent mechanism. Environ Health Perspect 120 984 989, doi:10.1289/ehp.1205063 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

