Quantitative analysis of intrinsic and extrinsic factors in the aggregation mechanism of Alzheimer-associated Aβ-peptide
- PMID: 26758487
- PMCID: PMC4725935
- DOI: 10.1038/srep18728
Quantitative analysis of intrinsic and extrinsic factors in the aggregation mechanism of Alzheimer-associated Aβ-peptide
Abstract
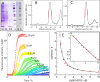
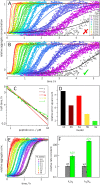
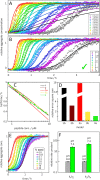
Disease related mutations and environmental factors are key determinants of the aggregation mechanism of the amyloid-β peptide implicated in Alzheimer's disease. Here we present an approach to investigate these factors through acquisition of highly reproducible data and global kinetic analysis to determine the mechanistic influence of intrinsic and extrinsic factors on the Aβ aggregation network. This allows us to translate the shift in macroscopic aggregation behaviour into effects on the individual underlying microscopic steps. We apply this work-flow to the disease-associated Aβ42-A2V variant, and to a variation in pH as examples of an intrinsic and an extrinsic perturbation. In both cases, our data reveal a shift towards a mechanism in which a larger fraction of the reactive flux goes via a pathway that generates potentially toxic oligomeric species in a fibril-catalyzed reaction. This is in agreement with the finding that Aβ42-A2V leads to early-onset Alzheimer's disease and enhances neurotoxicity.
Figures


References
-
- Glenner G. G. & Wong C. W. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890 (1984). - PubMed
-
- Hardy J. & Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol Sci. 12, 383–388 (1991). - PubMed
-
- Knowles T. P., Vendruscolo M. & Dobson C. M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell. Biol. 15, 384–396 (2014). - PubMed
-
- Walsh D. M. et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539 (2002). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

