4D cone-beam CT reconstruction using multi-organ meshes for sliding motion modeling
- PMID: 26758496
- PMCID: PMC5026392
- DOI: 10.1088/0031-9155/61/3/996
4D cone-beam CT reconstruction using multi-organ meshes for sliding motion modeling
Abstract
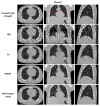
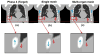
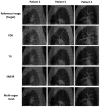
A simultaneous motion estimation and image reconstruction (SMEIR) strategy was proposed for 4D cone-beam CT (4D-CBCT) reconstruction and showed excellent results in both phantom and lung cancer patient studies. In the original SMEIR algorithm, the deformation vector field (DVF) was defined on voxel grid and estimated by enforcing a global smoothness regularization term on the motion fields. The objective of this work is to improve the computation efficiency and motion estimation accuracy of SMEIR for 4D-CBCT through developing a multi-organ meshing model. Feature-based adaptive meshes were generated to reduce the number of unknowns in the DVF estimation and accurately capture the organ shapes and motion. Additionally, the discontinuity in the motion fields between different organs during respiration was explicitly considered in the multi-organ mesh model. This will help with the accurate visualization and motion estimation of the tumor on the organ boundaries in 4D-CBCT. To further improve the computational efficiency, a GPU-based parallel implementation was designed. The performance of the proposed algorithm was evaluated on a synthetic sliding motion phantom, a 4D NCAT phantom, and four lung cancer patients. The proposed multi-organ mesh based strategy outperformed the conventional Feldkamp-Davis-Kress, iterative total variation minimization, original SMEIR and single meshing method based on both qualitative and quantitative evaluations.
Figures















References
-
- Agostoni E, Zocchi L. Mechanical coupling and liquid exchanges in the pleural space. Clin Chest Med. 1998;19:241–60. - PubMed
-
- Ahn B, Kim J. Measurement and characterization of soft tissue behavior with surface deformation and force response under large deformations. Med Image Anal. 2010;14:138–48. - PubMed
-
- Berg MD, Cheong O, Kreveld MV, Overmars M. Computational Geometry: Algorithms and Applications. Berlin: Springer; 2008.
-
- Bergner F, Berkus T, Oelhafen M, Kunz P, Pa T, Grimmer R, Ritschl L, Kachelriess M. An investigation of 4D cone-beam CT algorithms for slowly rotating scanners. Med Phys. 2010;37:5044–53. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
