EGFR Signaling Enhances Aerobic Glycolysis in Triple-Negative Breast Cancer Cells to Promote Tumor Growth and Immune Escape
- PMID: 26759242
- PMCID: PMC4775355
- DOI: 10.1158/0008-5472.CAN-15-2478
EGFR Signaling Enhances Aerobic Glycolysis in Triple-Negative Breast Cancer Cells to Promote Tumor Growth and Immune Escape
Abstract
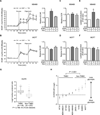
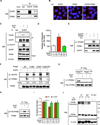
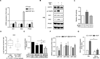
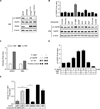
Oncogenic signaling reprograms cancer cell metabolism to augment the production of glycolytic metabolites in favor of tumor growth. The ability of cancer cells to evade immunosurveillance and the role of metabolic regulators in T-cell functions suggest that oncogene-induced metabolic reprogramming may be linked to immune escape. EGF signaling, frequently dysregulated in triple-negative breast cancer (TNBC), is also associated with increased glycolysis. Here, we demonstrated in TNBC cells that EGF signaling activates the first step in glycolysis, but impedes the last step, leading to an accumulation of metabolic intermediates in this pathway. Furthermore, we showed that one of these intermediates, fructose 1,6 bisphosphate (F1,6BP), directly binds to and enhances the activity of the EGFR, thereby increasing lactate excretion, which leads to inhibition of local cytotoxic T-cell activity. Notably, combining the glycolysis inhibitor 2-deoxy-d-glucose with the EGFR inhibitor gefitinib effectively suppressed TNBC cell proliferation and tumor growth. Our results illustrate how jointly targeting the EGFR/F1,6BP signaling axis may offer an immediately applicable therapeutic strategy to treat TNBC.
©2016 American Association for Cancer Research.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures


References
-
- Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

