Identification of DNA Methylation-Independent Epigenetic Events Underlying Clear Cell Renal Cell Carcinoma
- PMID: 26759245
- PMCID: PMC4873378
- DOI: 10.1158/0008-5472.CAN-15-2622
Identification of DNA Methylation-Independent Epigenetic Events Underlying Clear Cell Renal Cell Carcinoma
Abstract
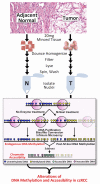
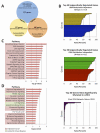
Alterations in chromatin accessibility independent of DNA methylation can affect cancer-related gene expression, but are often overlooked in conventional epigenomic profiling approaches. In this study, we describe a cost-effective and computationally simple assay called AcceSssIble to simultaneously interrogate DNA methylation and chromatin accessibility alterations in primary human clear cell renal cell carcinomas (ccRCC). Our study revealed significant perturbations to the ccRCC epigenome and identified gene expression changes that were specifically attributed to the chromatin accessibility status whether or not DNA methylation was involved. Compared with commonly mutated genes in ccRCC, such as the von Hippel-Lindau (VHL) tumor suppressor, the genes identified by AcceSssIble comprised distinct pathways and more frequently underwent epigenetic changes, suggesting that genetic and epigenetic alterations could be independent events in ccRCC. Specifically, we found unique DNA methylation-independent promoter accessibility alterations in pathways mimicking VHL deficiency. Overall, this study provides a novel approach for identifying new epigenetic-based therapeutic targets, previously undetectable by DNA methylation studies alone, that may complement current genetic-based treatment strategies. Cancer Res; 76(7); 1954-64. ©2016 AACR.
©2016 American Association for Cancer Research.
Figures




References
-
- Gupta KM, J. D, Li JZ, Russell MW, Charbonneau C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): A literature review. Cancer Treatment Reviews. 2008;34(3):193–205. - PubMed
-
- Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–32. - PubMed
-
- Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol. 2014;11(9):517–25. - PubMed
-
- Dancey JE, Bedard PL, Onetto N, Hudson TJ. The genetic basis for cancer treatment decisions. Cell. 2012;148(3):409–20. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

