Cross-backbone templating; ribodinucleotides made on poly(C)
- PMID: 26759450
- PMCID: PMC4748817
- DOI: 10.1261/rna.054866.115
Cross-backbone templating; ribodinucleotides made on poly(C)
Abstract
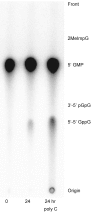
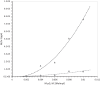
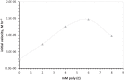
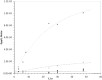
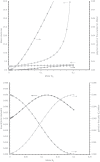
G(5')pp(5')G synthesis from pG and chemically activated 2MeImpG is accelerated by the addition of complementary poly(C), but affected only slightly by poly(G) and not at all by poly(U) and poly(A). This suggests that 3'-5' poly(C) is a template for uncatalyzed synthesis of 5'-5' GppG, as was poly(U) for AppA synthesis, previously. The reaction occurs at 50 mM mono- and divalent ion concentrations, at moderate temperatures, and near pH 7. The reactive complex at the site of enhanced synthesis of 5'-5' GppG seems to contain a single pG, a single phosphate-activated nucleotide 2 MeImpG, and a single strand of poly(C). Most likely this structure is base-paired, as the poly(C)-enhanced reaction is completely disrupted between 30 and 37 °C, whereas slower, untemplated synthesis of GppG accelerates. More specifically, the reactive center acts as would be expected for short, isolated G nucleotide stacks expanded and ordered by added poly(C). For example, poly(C)-mediated GppG production is very nonlinear in overall nucleotide concentration. Uncatalyzed NppN synthesis is now known for two polymers and their complementary free nucleotides. These data suggest that varied, simple, primordial 3'-5' RNA sequences could express a specific chemical phenotype by encoding synthesis of complementary, reactive, coenzyme-like 5'-5' ribodinucleotides.
Keywords: RNA gene; coenzyme; cofactor; phenotype; template.
© 2016 Majerfeld et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





References
-
- Bochner BR, Ames BN. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem 257: 9759–9769. - PubMed
-
- Cafferty BJ, Gállego I, Chen MC, Farley KI, Eritja R, Hud NV. 2013. Efficient self-assembly in water of long noncovalent polymers by nucleobase analogues. J Am Chem Soc 135: 2447–2450. - PubMed
-
- Ferris JP, Hill JAR, Liu R, Orgel LE. 1996. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 381: 59–61. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
