Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast
- PMID: 26759455
- PMCID: PMC4748822
- DOI: 10.1261/rna.054452.115
Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast
Abstract
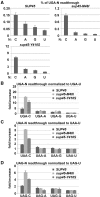
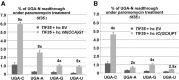
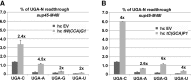
The molecular mechanism of stop codon recognition by the release factor eRF1 in complex with eRF3 has been described in great detail; however, our understanding of what determines the difference in termination efficiencies among various stop codon tetranucleotides and how near-cognate (nc) tRNAs recode stop codons during programmed readthrough in Saccharomyces cerevisiae is still poor. Here, we show that UGA-C as the only tetranucleotide of all four possible combinations dramatically exacerbated the readthrough phenotype of the stop codon recognition-deficient mutants in eRF1. Since the same is true also for UAA-C and UAG-C, we propose that the exceptionally high readthrough levels that all three stop codons display when followed by cytosine are partially caused by the compromised sampling ability of eRF1, which specifically senses cytosine at the +4 position. The difference in termination efficiencies among the remaining three UGA-N tetranucleotides is then given by their varying preferences for nc-tRNAs. In particular, UGA-A allows increased incorporation of Trp-tRNA whereas UGA-G and UGA-C favor Cys-tRNA. Our findings thus expand the repertoire of general decoding rules by showing that the +4 base determines the preferred selection of nc-tRNAs and, in the case of cytosine, it also genetically interacts with eRF1. Finally, using an example of the GCN4 translational control governed by four short uORFs, we also show how the evolution of this mechanism dealt with undesirable readthrough on those uORFs that serve as the key translation reinitiation promoting features of the GCN4 regulation, as both of these otherwise counteracting activities, readthrough versus reinitiation, are mediated by eIF3.
Keywords: GCN4; eRF1; programmed stop codon readthrough; termination; tetranucleotide; uORF.
© 2016 Beznosková et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
-
- Bidou L, Allamand V, Rousset JP, Namy O. 2012. Sense from nonsense: therapies for premature stop codon diseases. Trends Mol Med 18: 679–688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
