Super-resolution imaging reveals distinct chromatin folding for different epigenetic states
- PMID: 26760202
- PMCID: PMC4905822
- DOI: 10.1038/nature16496
Super-resolution imaging reveals distinct chromatin folding for different epigenetic states
Abstract
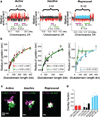
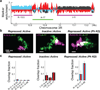
Metazoan genomes are spatially organized at multiple scales, from packaging of DNA around individual nucleosomes to segregation of whole chromosomes into distinct territories. At the intermediate scale of kilobases to megabases, which encompasses the sizes of genes, gene clusters and regulatory domains, the three-dimensional (3D) organization of DNA is implicated in multiple gene regulatory mechanisms, but understanding this organization remains a challenge. At this scale, the genome is partitioned into domains of different epigenetic states that are essential for regulating gene expression. Here we investigate the 3D organization of chromatin in different epigenetic states using super-resolution imaging. We classified genomic domains in Drosophila cells into transcriptionally active, inactive or Polycomb-repressed states, and observed distinct chromatin organizations for each state. All three types of chromatin domains exhibit power-law scaling between their physical sizes in 3D and their domain lengths, but each type has a distinct scaling exponent. Polycomb-repressed domains show the densest packing and most intriguing chromatin folding behaviour, in which chromatin packing density increases with domain length. Distinct from the self-similar organization displayed by transcriptionally active and inactive chromatin, the Polycomb-repressed domains are characterized by a high degree of chromatin intermixing within the domain. Moreover, compared to inactive domains, Polycomb-repressed domains spatially exclude neighbouring active chromatin to a much stronger degree. Computational modelling and knockdown experiments suggest that reversible chromatin interactions mediated by Polycomb-group proteins play an important role in these unique packaging properties of the repressed chromatin. Taken together, our super-resolution images reveal distinct chromatin packaging for different epigenetic states at the kilobase-to-megabase scale, a length scale that is directly relevant to genome regulation.
Figures
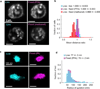




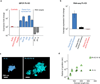
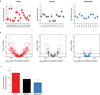



Comment in
-
Painting a Clearer Picture of Chromatin.Dev Cell. 2016 Feb 22;36(4):356-7. doi: 10.1016/j.devcel.2016.02.002. Dev Cell. 2016. PMID: 26906730 Free PMC article.
References
-
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. - PubMed
-
- Bickmore WA. The Spatial Organization of the Human Genome. Annu. Rev. Genomics Hum. Genet. 2013:67–84. at < http://www.ncbi.nlm.nih.gov/pubmed/23875797>. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

